Research Article - (2025) Volume 16, Issue 1
A Bacteriological Profile of Neonatal Infections at the Charles De Gaulle Pediatric University Hospital in Ouagadougou (Burkina Faso) from 2019 to 2023
Absatou BA/KY1*,
Issa Tonde2,
Marcel Sawadogo3,
Thierry Djiguemde2,
Arnaud Diendere1,
Idriss Traore1,
Coulibaly Drissa2,
Salimata Diallo1,
Issoufou Ouedraogo1 and
Idrissa Sanou4
1Department of Pediatrics, Bogodogo University Hospital, Ouagadougou, Burkina Faso
2Department of Pediatrics, Charles De Gaulle University Pediatric Hospital, Ouagadougou, Burkina Faso
3Department of Pediatrics, Yalgado Ouedraogo University Hospital Center, Ouagadougou, Burkina Faso
4Department of Pediatrics, Tengadogo University Hospital Center, Ouagadougou, Burkina Faso
*Correspondence:
Absatou BA/KY, Department of Pediatrics, Bogodogo University Hospital, Ouagadougou,
Burkina Faso,
Email:
Received: 05-May-2025, Manuscript No. IPACM-25-15709;
Editor assigned: 12-May-2025, Pre QC No. IPACM-25-15709;
Reviewed: 26-May-2025, QC No. IPACM-25-15709;
Revised: 20-Aug-2025, Manuscript No. IPACM-25-15709;
Published:
18-Sep-2025
Abstract
Introduction: Neonatal infection is a public health problem due to its frequency and severity. Probabilistic antibiotic therapy must be as effective as possible, hence the importance of knowing the bacteriological profile in order to improve the care of newborns. The objective of this study is to describe the bacteriological profile of neonatal infections in the neonatology unit of CHUP-CDG.
Materials and methods: This was a retrospective, descriptive cross-sectional study over a 5-year period, from January 1, 2019, to December 31, 2023. It included all samples from newborns in the neonatology unit who underwent cytobacteriological examination at the CHUP-CDG laboratory. All samples taken for suspected neonatal infection were included in the study, the results of which were duly reported in the registry during the study period.
Results: A total of 246 biological samples were collected, of which 98 (39.8%) were culture-positive. Klebsiella pneumoniae, Staphylococcus aureus and Escherichia coli were the most frequently identified bacterial species. As for sensitivity, it was good against vancomycin, amikacin, the combination of piperacillin and tazobactam, imipenem, netilmycin and chloramphenicol. High resistance to penicillins (penicillin G, ampicillin, ticarcillin), cephalosporins (cefalotin, ceftriaxone and cefepime), also macrolides (azithromycin) and cyclines (tetracycline).
Conclusion: The evolution of resistance phenotypes requires the rational use of antibiotics.
Keywords
Neonatal infections; Bacterial strains; Resistance phenotype; Antibiotic susceptibility
Introduction
The neonatal period is the stage of life when humans are most fragile, therefore vulnerable to several diseases, including infections. Neonatal infection is a clinical syndrome of bacteraemia characterized by clinical signs and symptoms occurring in a newborn [1]. Neonatal Infection (NNI) is a public health problem because of its frequency and severity [2]. It is a major concern in both developing and industrialized countries [3]. According to the WHO, the global mortality rate was 18 per 1,000 in 2021 [4].
Africa is the most affected continent where the neonatal mortality rate is 28.0 per 1,000 live births, followed by the Eastern Mediterranean (10.6) and Southeast Asia (24.3) [5]. Sub-Saharan Africa has the highest infant mortality rates in the world, with 74 deaths per 1,000 live births [6]. There are an estimated 6.9 million annual serious neonatal bacterial infections needing treatment and one-third occur in sub-Saharan Africa, despite evidence of 25% reductions in all-cause neonatal mortality with immediate antibiotic treatment [7].
In Burkina Faso, this rate is 26 per 1000 live births [8]. Newborn deaths account for 41% of deaths in children under 5 years of age [9]. The microbiological profile of infections varies across geographic regions and evolves over time, requiring adaptation of initial probabilistic antibiotic therapy to bacterial ecology [10]. Geographic differences in microbiology are likely related to a diverse prevalence of maternal and neonatal risk factors such as prematurity, obstetric and neonatal care practices and regional variations in community flora [11].
Efforts are being made globally to reduce neonatal mortality. In this regard, in 2006, Burkina Faso launched a policy to improve emergency obstetric and neonatal care in collaboration with the World Health Organization (WHO). This policy consisted in creating an environment conducive to care, strengthening the capacities of providers and subsidizing care [12]. This led to the implementation of the policy of free care for pregnant women and children under five in 2016 in order to strengthen families' access to care [13]. In addition to these actions, the Ministry of Health has developed modules, provided training and established audits of university hospitals and regional hospitals to assess the causes of maternal and neonatal deaths [14]
Although these efforts have contributed to reducing the mortality rate, it remains a concern in Burkina Faso with difficulties in early diagnosis and the emergence of multi-resistant bacteria in infections. The treatment and prognosis of neonatal infections are based on rapid and effective antibiotic therapy, generally probabilistic in the first 48 hours and then based on the identification of the main species encountered and their sensitivity to antibiotics.
Probabilistic antibiotic therapy remains necessary and must be as effective as possible, hence the importance of knowing the epidemiological profile of germs encountered in pathological products in order to establish their sensitivity to antibiotics and better guide antibiotic therapy [15]. But also prenatal monitoring is essential to assess the health of the mother and the fetus, in particular by screening for certain infections that can have serious consequences on the baby's development. The acronym TORCH groups together several important prenatal infections to monitor, including toxoplasmosis, rubella, cytomegalovirus and herpes simplex virus [16]. It is in this context that this study was initiated to describe the bacteriological profile of neonatal infections in the neonatology unit of the Charles-De-Gaulle Pediatric University Hospital (CDG - PUH) with the aim of improving the care of newborns
Materials and Methods
Study site
The study was conducted at the CHUP-CDG in Ouagadougou. It is a referral hospital for third-level pediatric care in the national health pyramid.
Study type and period
This was a descriptive cross-sectional study with retrospective data collection over a 5-year period, from January 1, 2019, to December 31, 2023.
Study population
The study included all newborn samples from the neonatology unit for whom cytobacteriological examination was performed in the CDG-PUH laboratory. All newborn samples identified a bacterial strain through cytobacteriological examination in the CDG-PUH laboratory.
Bacteriological analysis
Pus was collected using two sterile swabs or in a test tube when abundant. One swab was used for microscopy and the other for isolation. For blood culture, the sample was directly inoculated into the appropriate culture broths and brought to the laboratory. The volume of blood collected from the infant was 1 mL-3 mL. The perineal region was disinfected with soap and an adhesive bag (Urinocol) was attached to collect the urine.
The macroscopic examination consisted of assessing the appearance, consistency and color of the pathological products. The microscopic examination was carried out in two stages: qualitative microscopy allowed to assess the morphology, mobility, abundance of bacteria and also the presence of other elements such as: Leukocytes, yeasts and red blood cells, crystals, etc. and quantitative microscopy which concerned the Cerebrospinal Fluid (CSF), urine, allowed to count the number of leukocytes, red blood cells. For each sample, a Gram-stained smear was read under the microscope. The samples were then cultured in the appropriate culture media. For urine, a drop (10 uL) was taken and placed on the surface of the medium: Cystine-Lactose-Electrolyte-Deficient (CLED) and a depletion in tight streaks in a single dial was carried out. For other pathological products: exhaustion in tight streaks 3 dials were produced.
The bacteria were identified based on their morphological, cultural and biochemical characteristics.
The antibiogram was performed using the Kirby Bauer technique or the agar diffusion technique, as recommended by CASFM/Eucast 22 and the national antibiogram guide.
Resistance phenotype investigation
Detection of Extended Spectrum Beta-Lactamases (ESBL): After inoculation of a suspension on Muller Hinton (MH) agar, the production of ESBL is observed after performing the double diffusion technique between a disc of Amoxicillin+clavulanic acid (AMC) or Ticarcillin+Clavulanic acid (TTC) and discs of 3rd generation (C3G) or 4th generation (C4G) cephalosporins: Ceftriaxone, Ceftazidime, Cefepime, Cefixime at a distance of 20 mm-30 mm. The production of ESBL is revealed by the presence of synergy between C3G or C4G and the inhibitor at 37°C after 24 hours of incubation, the result is said to be positive if a characteristic image called "champagne cork" appears. The synergy test is said to be negative if there is no “champagne cork” image after bringing the antibiotic discs together.
Determination of other phenotypes (penicillinase when S. aureus is resistant to penicillin G and MRSA if S. aureus is resistant to cefoxitin).
Processing and analysis of collected data
The Excel file was imported into EPI-Info version 7.2.6.0 for data analysis by determining frequencies, cross-referencing parameters, etc. The results were expressed in tables and figures generated by Microsoft Excel version 2019.
Processing and analysis of collected data
The excel file was imported into EPI-Info version 7.2.6.0 for data analysis by determining frequencies, cross referencing parameters, etc. The results were expressed in tables and figures generated by Microsoft Excel version 2019.
Results
A total of 246 biological samples from the neonatology unit, consisting of pus and serosum, blood and urine, were received at the bacteriology virology unit of the laboratory during the study period. Analysis of these samples identified 98 bacterial strains, distributed among the different biological products, representing a positivity rate of 39.8%, as shown in Table 1.
|
Nature
|
Number
|
Positive culture
|
Percentage (%)
|
|
Pus and serosity
|
132
|
67
|
50.75
|
|
Blood (haemoculture)
|
96
|
25
|
26.04
|
|
Urine
|
18
|
6
|
33.33
|
|
Total
|
246
|
98
|
39.83
|
Table 1: Positivity rates of different biological samples.
Sociodemographic characteristics
A total of 60 positive cultures (61.2%) came from female newborns, representing a sex ratio of 0.63. The mean age of newborns with positive cultures was 7.9 days, with a range of 0 to 28 days and a standard deviation of 8.191. The 0-3-day age group was the most represented (41.8%). The late neonatal period accounted for 58.2% (Figure 1).
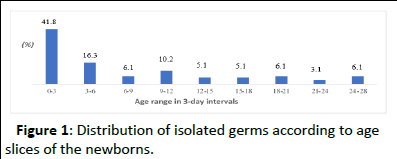
Figure 1: Distribution of isolated germs according to age slices of the newborns.
Bacteriological profile
Among the 98 bacterial strains isolated, Klebsiella pneumoniae, Staphylococcus aureus and Escherichia coli were the most frequently identified bacterial species, accounting for 29.60%, 14.30% and 13.30%, respectively, as shown in Table 2.
| Isolated germ |
Number |
Percentage |
| Klebsiella pneumoniae |
29 |
29.6 |
| Staphylococcus aureus |
14 |
14.3 |
| Escherichia coli |
13 |
13.3 |
| Pseudomonas aeruginosa |
9 |
9.2 |
| Staphylococcus saprophyticus |
9 |
9.2 |
| Klebsiella oxytoca |
6 |
6.1 |
| Candida sp |
4 |
4.1 |
| Enterobacter sp |
4 |
4.1 |
| Enterobacter cloacae |
3 |
3.1 |
| Streptococcus sp |
3 |
3.1 |
| Citrobacter jneuindii |
1 |
1 |
| Acinetobacter baumannii |
1 |
1 |
| Stenotrophomonas maltophilia |
1 |
1 |
| Serratia odorifera |
1 |
1 |
| Total |
98 |
1 |
Table 2: Specific frequency of bacterial strains isolated from newborns in the neonatology unit of the CDG-PUH between 2019 and 2023.
Antibiotic sensitivities
The 35 Klebsiella species strains isolated had good sensitivity to imipenem (82%) and amikacin (92%). High resistance was observed with beta-lactam antibiotics, particularly amoxicillin+clavulanic acid (86%), ticarcillin (100%), piperacillin (87%), C3Gs such as cefepime (88%), ceftriaxone (95%), ceftazidine (81%) and aminoglycosides such as gentamicin (89%) and netilmycin (80%). Finally, cotrimoxazole (88%). Compared to the resistance phenotype of Klebsiella sp, Table 3 shows 18 ESBL-producing strains or 51.43%.
| Phenotypes |
Number |
Percentage |
| Savage |
4 |
44.44 |
| Efflux |
2 |
22.22 |
| Loss of porin D2 |
2 |
22.22 |
| Cephalosporinase |
1 |
11.11 |
| Total |
9 |
1 |
Table 3: Resistance phenotype of Klebsiella sp strains.
Among the 23 Staphylococcus sp strains isolated, their susceptibility was good to netilmycin (100%), clindamycin (100%), thiamphenicol/chloramphenicol (100%), amoxicillin+clavulanic acid (83%), ceftriaxone (87%) and cefoxitin (75%). High resistance was observed with penicillin (89%), erythromycin (61%).
Among the resistance phenotypes of Staphylococcus strains, 17 (74%) were penicillinase-producing and 22% were MRSA.
Regarding the susceptibility profile of the 13 Escherichia coli strains isolated, imipenem and chloramphenicol (100%) and amikacin (70%) were susceptible. High resistance was observed with amoxicillin+clavulanic acid (70%), ampicillin (75%), ceftriaxone (83%), cefepime (71%) and cotrimoxazole (70%).
Among the resistance phenotypes, 7 (54%) E. coli strains developed the ESBL phenotype followed by cephalosporinase (38.4%) and one wild-type strain (8%).
The susceptibility profile of the 9 Pseudomonas sp strains was good against piperacillin+tazobactam (100%), gentamicin (100%), ceftazidine (83.33%) and ciprofloxacin (80%). Resistance was 75% for cotrimoxazole, 50% for aztreonam and 100% (amoxicillin+clavulanic acid). Regarding the resistance phenotypes for this germ: Wild-type strains represented 44% or 4 strains, as shown in Table 3. Resistance phenotype of Pseudomonas sp strains isolated from newborns, neonatology unit of CDG-PUH between 2019-2023.
Discussion
During our study period, 246 biological samples from suspected cases of neonatal infections underwent cytobacteriological examination. The prevalence of confirmed neonatal bacterial infections was 39.8%, which is consistent with data from the literature, which is 30 to 40%. Savadogo, et al. in Burkina Faso in 2010 [17] found a 28.25% bacteriological examination confirmation rate in a study on suspected neonatal infections. This result could be explained by improved identification techniques and increased hospital attendance due to free care at the neonatal unit of the CDG-PUH.
Females were more prevalent (61.2%) in the total population. The sex ratio was 0.63 in favor of girls. Sawadogo, et al, in Burkina 2010 [17] observed a sex ratio of 0.7 in a study conducted at CHUP-CDG on urinary tract infection in infants. The high frequency in females could be explained by the significant proportion of females in the general population according to the demographic survey carried out in Burkina Faso [18].
The average age of newborns in our series was 7.9 days with extremes of 0 days and 28 days. The most affected age group was between the 4th and 28th day or 58.2%. These results are comparable to those of Kabre, et al, in 2015[19] at CHUP-CDG who observed 50.4% of this age group. This could be explained by postnatal bacterial contaminations which are most often nosocomial infections. These infections are frequent in neonatology due to the immaturity of the immune system of newborns, massive contamination from the environment, staff, duration) [20]. Efforts on staff training and strengthening the technical platform of care and hospital hygiene measures must be taken into account when implementing neonatal infection control measures at CDG-PUH. Neonatal sepsis is a clinical syndrome caused by severe systemic infection during the first month of life.
In our study, sepsis accounted for 25.5% of laboratory confirmed neonatal infections. The high rate of this infection could be related to vertical transmission of pathogens across the placenta or during delivery from the maternal genitourinary tract, but also to direct exposure of the newborn at birth or to prolonged catheterization in the bloodstream. The majority of identified strains came from pus and serosity samples, which included body fluids, joint fluids, hematomas, abscesses and superficial skin and tissue lesions 67 samples or (68.4%), followed by blood 25 samples or (25.5%) and urine 6 samples or (6.1%). These results are similar to those of Kabre et al., who observed in 2015 at the CDG-PUH that 53.34% of samples were composed of pus and serosity.
It should be noted that 2021 was the year that recorded more than 35 (35.7%) cases of neonatal infections. This can be attributed to certain pathogens such as the dengue virus and SARcoV2 which weaken the newborn and expose them to bacterial superinfections, which could explain the increase in hospitalizations observed between 2020 and 2021. Regarding the isolates in the present study, it was revealed that Klebsiella pneumoniae (29.6%), Staphylococcus aureus (14.3%) and Escherichia coli (13.3%) were the most frequently identified bacterial species, however, Dao's team [21] in 2012 observed (38%) Escherichia coli, (15%) Klebsiella pneumoniae and (13.4%) Staphylococcus aureus. In 2014, in the same hospital, another study revealed that the most frequently identified germs were Escherichia coli, Enterococcus sp, Pseudomonas sp and Staphylococcus aureus, with a constant frequency of 13.3%.
According to a study conducted by ZAIDI et al. [22] on pathogens linked to sepsis in newborns and young infants in developing countries, Klebsiella species (25%), Escherichia coli (15%) and Staphylococcus aureus (18%) were the main pathogens incriminated. The identification of these hospital-associated germs could be explained by the inadequacy of incubators, the frequent use of intensive care with the use of invasive equipment, immature neonatal immunity and the geographical variation of causative germs.
In blood cultures, Klebsiella pneumoniae was the most isolated germ with a frequency of 40%. This result corroborates that of Kamaye, et al [23] in Niger in 2019, which found 32.8%. The relatively high incidence of Klebsiella pneumoniae may be due to the simultaneous presence of nosocomial infections in the neonatal unit during the period of our study. The high incidence of Staphylococcus aureus infections reflects the inadequacy of hygiene measures in hospitals. Poor aseptic conditions and a shortage of nursing staff may partly explain this situation.
Klebsiella sp strains were more resistant than susceptible, with higher susceptibility to imipenem (82%) and amikacin (92%). Kemeze, et al. [24] observed susceptibility rates similar to ours in Douala in 2016.
Resistance was very high with amoxicillin and clavulanic acid (86%), ticarcillin (100%), piperacillin (87%), netilmycin (80%), cephalosporins (cefepime 88%, ceftriaxone 95%, ceftazidine 81%); and cotrimoxazole (89%). In Nepal, Pokhrel et al. [25] observed that Klebsiella sp was highly resistant to commonly used antibiotics such as cefotaxime (90.5%), gentamicin (75%), ciprofloxacin (76.2%), ofloxacin (72.2%) and chloramphenicol (65%). Jemal et al., found that the least effective antimicrobials against Klebsiella sp were ampicillin, amoxicillin, gentamicin and ceftriaxone, with a resistance rate of 100%, 96%, 72% and 90% respectively. These results could be attributed to overuse of antibiotics, as well as to hyperproduction of chromosomal cephalosporinases or to the acquisition of genes producing extended-spectrum beta lactamases (ESBL at 51.43%, cephalosporinase at 28.57%).
Staphylococcus sp strains were more susceptible than resistant. Susceptibility to antibiotics such as netilmycin (100%), oxacillin (83%), amoxicillin clavulanic acid (87%), ceftriaxone (87%), thiamphenicol/chloramphenicol (100%), clindamycin (100%) and tobramycin (100%) was good. According to a study conducted in a tertiary hospital in Nigeria, Staphylococcus sp was found to have susceptibility to amoxicillin-clavulanic acid 89%, cefuroxime 85%, ciprofloxacin 75%, chloramphenicol 71% and erythromycin 64%. This could be due to the decrease or loss of acquired resistance mechanisms. Escherichia coli strains were more resistant than susceptible with resistance observed to ceftriaxone 83%, cefepime 71%, amoxicillin/clavulanic acid 70%, ampicillin 75% and cotrimoxazole 70%. The level of resistance to ampicillin (96%) and ceftriaxone (67%) was also observed by Jemal, et al. This result could be due to inappropriate or excessive use of antibiotics which resulted in the development of BLES 53.87% and cephalosporinase 38.46% resistance phenotypes.
Pseudomonas sp strains were more susceptible than resistant with susceptibility to ceftazidine 83.33%, gentamicin 100%, piperacillin+tazobactam 100%, piperacillin 100%, ciprofloxacin 80% and amikacin 100%. Pseudomonas showed 100% resistance to ceftriaxone in our study. The reduced susceptibility to β-lactams could be due to the emergence and development of resistance phenotypes. Imipenem, amikacin and piperacillin+tazobactam, which are effective antibiotics, are difficult to obtain and access for patients due to their high price and their route of administration which is essentially parenteral for amikacins. It is therefore essential to rationalize the use of antibiotics in order to control this antibiotic resistance, while strengthening hygiene measures in healthcare facilities. It is also important for the clinician to have a precise approach to probabilistic treatments by using the most active antibiotics provided by the laboratory.
Conclusion
This study revealed that the majority of isolated organisms originated from pus and serosal samples. Late infections were the most common and female newborns were the most affected in our study. The bacteriological profile established in this study indicates that the isolates consisted mainly of Enterobacteriaceae, including the genera Klebsiella, Escherichia and Staphylococcus.
Regarding resistance, the identified organisms were more resistant than susceptible, with worrisome resistance to beta-lactam antibiotics (ampicillin, amoxicillin/clavulanic acid, ticarcillin, etc) and third-generation cephalosporins (ceftriaxone, cefepime, etc.). The emergence and evolution of resistance phenotypes such as penicillinase, cephalosporinase, ESBL and MRSA were noted. The origin of these pathogens needs to be further explored to take corrective measures to ensure good overall care of newborns.
Ethical Considerations
The study was conducted in an academic setting and the study protocol was submitted for approval to the General Management of the CDG - PUH (Children's Hospital Center) in order to obtain authorization to conduct the study. The confidentiality of patients' personal data was maintained during data collection. The collected data is coded to ensure anonymity and stored in a secure Excel database with a password to restrict access.
Conflicts of Interest
The author declares no conflicts of interest.
Author Contributions
Coulibaly Drissa: Wrote the body of the article. Absatou BA/KY: Principal Investigator and edited the article. Tondé Issa: Edited the article. Marcel Sawadogo: Edited the article. Thierry Djiguemdé: Edited the article. Arnaud Diendere: Edited the article. Traore Idriss: Edited the article. Diallo Salimata: Participated in data collection. Ouedraogo Issoufou: Participated in data collection. Sanou Idrissa: Edited the article.
Acknowledgments
The authors thank all the respondents who participated in this study. We also thank all those who provided essential material for the study.
References
- Gurdeep S D, Sonam A and Prateek S (2018) Etiological study of neonatal sepsis. Pediat Rev Int J Pediatr Res 5:19-23.
- Navon-Venezia S, Kondratyeva K, Carattoli A (2017) Klebsiella pneumoniae: A major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev 41:252-275.
[Crossref] [Google Scholar] [PubMed]
- Gras-Le Guen C, Launay E, Boscher C, Caillon J (2016) Update on neonatal infections. Bull National Acad Med 200:81-90.
- World Health Organization (2021). Neonatal mortality. Key facts UNICEF data.
- WHO (2015a) Data neonatal mortality. Global Health Observatory, World Health Organization, Geneva, Switzerland.
- Seale AC, Blencowe H, Manu AA, Nair H, Bahl R, et al. (2014) Estimates of potentially serious bacterial infections among newborns in sub-Saharan Africa, South Asia and Latin America in 2012: A systematic review and meta-analysis. Lancet Infect Dis 14:731-741.
- National Institutes of Health (NIH) (2023) Neonatal Mortal.
- WHO (2011) Neonatal mortality is declining too slowly, especially in Africa.
- Jemal M, Tinshku F, Nigussie Y, Kefyalew B, Alemu C, et al. (2021) Trend analysis of multidrugresistant bacterial pathogens causing neonatal seps is at University of Gondar comprehensive specialized Hospital, Northwest Ethiopia: A retrospective study. Int J Microbiol 21: 9992994
[Crossref] [Google Scholar] [PubMed]
- Mduma E, Halidou T, Kaboré B, Walongo T, Lompo P, et al. (2022) Etiology of severe invasive infections in young infants in rural settings in sub-Saharan Africa. PLoS One 17:e0264322
[Crossref] [Google Scholar] [PubMed]
- Zoungrana-Yameogo WN, Dahourou DL, Diallo AH, Sangho O, Nikiema E, et al. (2021) Neonatal mortality at Tengandogo University Hospital, Ouagadougou, Burkina Faso: A retrospective cohort study. J Interven Epidemiol Pub Health 4
[Google Scholar]
- The President of Burkina Faso (2016) DECREE 2016 311PRES/PM/MS/MATI)SI/MINEFID providing free healthcare for women and children under five living in Burkina Faso
- Ministry of Health. Establishment of audits of university hospitals and regional hospitals to assess the causes of maternal and neonatal deaths by the Ministry of Health.
- Mamadou T, Sylvie Z, Dissinviel K, Tani S, Rebeca T, et al. (2024) Bacteriological Profile of Effusion Fluids Infections at Charles De Gaulle University Pediatric Hospital from 2017 to 2020. Open J Med Microbiol 14:146-163
[Crossref] [Google Scholar]
- Apollo Hospitals (2025) Torch Syndrome: Causes, symptoms, diagnosis, treatment and prevention
- Savadogo H, Dao L, Tondé I, Tamini L, Ouédraogo AI, et al. (2021) Urinary tract infections in a pediatric setting: bacterial ecology and antibiotic susceptibility at the Pediatric Teaching Hospital Charles-de-Gaulle in Ouagadougou (Burkina Faso). Néphrol Ther 17:532-537
[Crossref] [Google Scholar] [PubMed]
- National Institute of Statistics and Demography (2019) Fifth General Population and Housing Census of Burkina Faso.
- Kabre R (2015) Bacteriological profile of neonatal infections and antibiotic sensitivity at the Charles de Gaulle Pediatric University Hospital in Ouagadougou. Pharmacy thesis. Ouagadougou: Joseph Ki Zerbo
- Rogombe SM, Jean K, Mimbila M, Kamgaing EK, M'ella RM (2018) Epidemiological Aspects and Evolution of Nosocomial Infection in the Neonatology Unit of Angondje Teaching Hospital. Neonat Pediatr Med 4:2
[Google Scholar]
- Dao L, Koueta F, Bationo R, Ouédraogo R, Ye D (2012) Neonatal bacterial infections in Ouagadougou: nature and sensitivity of germs. Sci Tech 35
- Zaidi AK, Thaver D, Ali SA, Khan TA (2009) Pathogens associated with sepsis in newborns and young infants in developing countries. Pediat Infec Dis J 28:10-18
[Crossref] [Google Scholar] [PubMed]
- Kamaye M, Alido S, Ibrahim D, Sani O, Aboubacar A (2019) Profile and sensitivity of bacteria isolated in early neonatal bacterial infections at the Issaka Gazobi maternity hospital in Niamey. J Afr Pediatr Genet Med 9:46-50
- Kemeze S, Moudze B, Chiabi A, Eposse C, Kaya A, et al. (2013) Clinical and bacteriological profile of neonatal bacterial infection at Laquintinie Hospital, Douala (Cameroon). Pan Afr Med J 23:97
[Crossref] [Google Scholar] [PubMed]
- Pokhrel B, Koirala T, Shah G, Joshi S, Baral P (2018) Bacteriological profile and antibiotic susceptibility of neonatal sepsis in neonatal intensive care unit of a tertiary hospital in Nepal. BMC Pediatr 18:208
[Crossref] [Google Scholar] [PubMed]
- Iregbu KC, Elegba OY, Babaniyi IB (2006) Bacteriological profile of neonatal septicaemia in a tertiary hospital in Nigeria. Afr Health Sci 6:151-154
[Crossref] [Google Scholar] [PubMed]
Citation: Absatou BA/KY, Tondé I, Sawadogo M, Djiguemdé T, Dienderé A, et al. (2025) A Bacteriological Profile of Neonatal Infections at the Charles
De Gaulle Pediatric University Hospital in Ouagadougou (Burkina Faso) from 2019 to 2023. Arch Clinic Microbiol Vol.16 No.1






