Keywords
Novel Coronavirus; SARS-CoV-2; Coronaviruses; Wuhan; China
Introduction
During the last few decades’ coronaviruses (CoVs) emerged as one of the most formidable public health challenges. Originally, reported in 1960s, human coronaviruses belong to the family Coronaviridae that is further divided into two subfamilies Orthocoronavirinaeand Torovirinae. Orthocoronavirinae consists of four genera, alpha, beta, gamma and delta coronaviruses. All four genera of coronaviruses can be found in the birds and mammals including bats. Avian infectious bronchitis viruses (IBV) were the first CoVs to be discovered which caused disease in chickens and humans. For example, both HCoV-229E and CoVOC43 were able to cause common cold in humans [1,2]. After the discovery of IBVs several other important CoVs such as SARS-CoV, MERS-CoV and SARS-CoV-2 were discovered which are associated with human infections [3,4]. However, the seventh human CoV (SARS-CoV-2) emerged as one of the most important CoVs, particularly because of the magnitude of outbreak and the rapid transmission of infections.
The name corona was derived from a Latin word, meaning “halo” or “crown” that was assigned because of its crown like projection. The virus is spherical in shape with a diameter of 120 nm-160 nm. Its genome consists of positive sense single stranded RNA (ssRNA+) ranging from 26 Kb-32 Kb in length [5]. The virus encodes four structural proteins namely M (membrane protein), E (envelope protein), S (spike protein) and N (nucleocapsid protein) [6]. The coronaviruses have been identified from different hosts including bats, camels, mice, masked palm civets, cats, dogs and from avian reservoir [7,8]. Except, sever acute respiratory syndrome (SARS-CoV) and Middle East respiratory syndrome (MERS-CoV), all other coronaviruses caused milder clinical symptoms in humans [7,8]. Only after an outbreak in the year 2002, a new coronavirus was identified, now known as sever acute respiratory syndrome (SARS-CoV) that caused severe infections in humans [9]. The SARS-CoV is a zoonotic virus and its first outbreak occurred in “ Guangdong ” province of People ’ s Republic of China. Sooner, the virus became a pandemic and more than 8000 human infections were recorded. Altogether, virus caused 774 deaths in 37 countries around the globe [10]. The palm civets were the natural reservoirs for the SARS-CoV [2]. The SARS-CoV infects both upper and lower respiratory tracts of humans. During the episode of infection, patient develops flu like symptoms, fever, chills, dry cough, breathlessness, and aches. In some cases, diarrhea that later progressed to acute respiratory distress syndrome was also reported [11]. In sever conditions, lung infection and pneumonia develop which leads to lung failure and even death [9]. Complications are more often reported in elderly and immune-compromised individuals. Likewise, other important coronavirus, MERS-CoV1 emerged in Arabian Peninsula and was first reported on June 13, year 2012 in Jeddah, Saudi Arabia [4]. The total numbers of laboratory confirmed cases were 2494, with 858 causalities and overall fatality rate was ~35% [8,12]. The reservoirs for the MERS-CoV were Arabian camels and its zoonotic transmission led to the lower respiratory tract infections in humans [13]. The MERS-CoV disseminated rapidly outside the Saudi Arabia. Due to this virus, in year 2015, the largest outbreak occurred in South Korea. The common symptoms included coughing, breathlessness, and fever. Though zoonotic in nature, MERSCoV disseminated rapidly through person to person contact. Taken together, until year 2002, coronaviruses were considered minor human pathogens and today they pose one of the most unprecedented challenges to human health prosperity, and social dynamics.
At the end of the year 2019, a novel coronavirus emerged in Wuhan, a city of People’s Republic of China. Initially identified as Sever Acute Respiratory Syndrome coronavirus-2 (SARSCoV- 2) it caused respiratory illness and pneumonia. On February 11, 2020, WHO announced for this novel coronavirus disease a standard format according to its nomenclatures Coronavirus Diseases-2019 (COVID-19) [14]. Subsequently, on the same day, this novel coronavirus was named as SARSCoV- 2 by the international committee on Taxonomy of Viruses (ICTV) [15]. Since the initial reports, more than n=6,267,488 laboratory and clinically confirmed cases (Update on 1st June 2020) have been reported which are expected to increase. The casualties are increasing on daily basis and at the time of compilation of this report total number of fatalities surpassed the figure n=373,961. However, among the total infected individuals n=2,847,571 (88%) recovered successfully whereas currently active cases are n=3,045,956 around the globe. Latest figures suggest that out of the total infected individuals n=2,992,556 (98%) are in less sever conditions, whereas n=53,400 (2%) are in critical conditions [16,17]. The virus spread very rapidly in USA and on June 1st, 2020, the laboratory confirmed cases were n=1,837,170 with total deaths n=106,195. The heights number of cases were reported from New York, New Jersey and lllinois (Total cases n=379,902; n=161,764 and n=120,260 respectively) and total deaths were (n=29,918; n=11,711 and n=5,390 respectively). In Pakistan, the index case was reported on 26th February 2020 and as of June 1st, 2020 the total confirmed cases were n=72,460 with total deaths n=1,543 [16]. Unfortunately, around the globe cases are continuing to increase consistently. On January 30, 2020, the World Health Organization (WHO) declared the outbreak of SARS-CoV-2 as a major public health emergency and a wider international concern. Till to date, Infections due to SARS-CoV-2 are reported from 213 different countries while on March 11, 2020 WHO declared it a global pandemic.
The index case of coronavirus was linked to local seafood market at Wuhan which sells live animals including bats, marmots, rabbits, and pangolin. However, recently diagnosed patients have no direct exposure to the market and are eventually the result of human to human transmission [18]. In terms of fatalities, so far, the most affected geographical areas are USA, UK, Italy, and Brazil. The highest numbers of casualties were reported from USA (n=106,195), followed by UK (n=38,489), Italy (n=33,415) and Brazil (n=29,341) [16]. At the initials stage, to reduce the spread of infections Government of People’s Republic of China took remarkable steps which were highly appreciated at international level and eventually led to the opening of Wuhan after 76 days of lockdown. Because of the strict measures including closing of schools, workplaces, cancelling flights and even entire cities were locked down in many countries around the globe [19]. In Iran majority of the cases were reported in the holy city of Qom, located in south of Tehran. From Iran spill over cases were reported in neighboring countries including Pakistan, Oman, Kuwait, Afghanistan, Iraq, and Bahrain. The spread of SARS-CoV-2 in South Korea was reported in a religious community in South Eastern city of Daegu. Highly vigilance policies and large-scale timely testing in South Korea significantly helped limiting the outbreak. Other countries like Spain, Romania, Denmark, and Algeria reported transmission from Italy [19]. Data from Robert Koch Institute confirmed n=183,494 cases in Germany. In short SARS-CoV-2 has been spreading with unprecedented pace and preventive measures seems to be less effective that stresses the need to learn fast and mount coordinated efforts to stop a second wave of pandemic. This report provides an overview of SARS-CoV-2 (Figure 1).
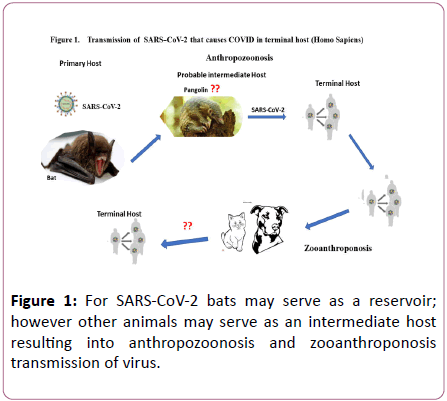
Figure 1: For SARS-CoV-2 bats may serve as a reservoir; however other animals may serve as an intermediate host resulting into anthropozoonosis and zooanthroponosis transmission of virus.
Structure and genome
Phylogenetic analysis and whole genome sequence of SARSCoV- 2, which is now the seventh human coronavirus was, elucidated recently [8]. The phylogenetic analysis confirmed that it belongs to the subgenus Sarbecovirus and shares similarity with coronavirus strains originating from bats. The virus SARS-CoV-2 shares 90% genetic similarity with bat-SLCoVZXC21, bat-SL-CoVZC45 rather to SARS-CoV or other human coronaviruses [8]. The current genomic data is based on the viral strains isolated from nine patients infected in Wuhan province of china (China National Microbiological Data Centre, accession number NMDC10013002, genomic accession number NMDC60013002-01 to NMDC60013002-10). It shows that all the sequenced strains shared 99.9% genetic similarity confirming common origin of SARS-CoV-2. Coronaviruses genomes have 10-4nucleotide substitution rate per site in a single year; hence this virus would mutate further and needs sequence-based surveillance around the globe. Based on the available data genetic recombination has been overruled for the emergence of SARS-CoV-2. Large numbers of full genome sequences are now available which use Wuhan-Hu-1/2019 as reference genome for comparison that would help us to understand genetic divergence of virus and dynamics of human transmission. The transmission capacity of the virus relies on envelop spike (S-protein) which plays crucial role in receptor mediated binding and membrane fusion [20]. In coronaviruses spike protein plays dual role, receptor mediated binding and membrane fusion which depends on functionality of S1 and S2 domain of spike protein [21]. The S2 protein of SARS-CoV-2shared 93% similarity with bat-SL-CoVZXC21, bat- SL-CoVZC45 strains while both N-terminal and C-terminal of S1 protein can facilitate receptor binding [22]. Interestingly, receptor binding domain located in S1 region of SARS-CoV-2 is closer to linage B and particularly SARS-CoV.
The spike protein is the main mediator of the host pathogen interaction and comprises of three main parts, S1 &S2 region (ectodomain), a transmembrane anchor and an intercellular tail [23]. It was shown that though S1 subunits of different coronavirus genera originated from a common ancestor over the period they attained evolutionary divergence. The RBD (receptor binding domain) located at the S1 region of ectodomain facilitates binding of the virion while the S2 region facilitates membrane fusion [24]. Overall, spike proteins of SARS-CoV-2 virus has 76% homology with Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV), while the RBD (193 resides) domain shares slightly less similarity that was because of mutations identified at the C-terminal region of the receptor binding domain [25,26]. Angiotensin-converting enzyme 2 (ACE2) serves as receptor for the SARS-CoV and interacts with S1 subunit of the spike protein from amino acid 318-510 and is required as minimal receptor binding domain. Since residues crucial for the receptor binding are intact, SARSCoV- 2 may bind effectively with human ACE2 receptor. The socalled receptor binding motif (RBM) that spans from amino acids 424–494, of the RBD establishes direct contact with ACE2 receptor. Recent modeling studies elucidated identical threedimensional orientations of RBD despite changes in these regions [26-28]. In fact, it is possible that elucidated mutations may even facilities stronger binding with human cell surface receptor because of favorable structural rearrangements, however, exact role of this region in SARS-CoV-2 binding remains to be elucidated [29]. Interestingly, in the region encoding spike protein of SARS-CoV-2, four insertion sequences having significant homology with gp120 (glycoprotein of HIV-1 also known as cellular receptor recognition protein) and Gag region of HIV-1 were identified [30]. Importantly, these sequences remain novel to only SARSCoV- 2 and raised concerns about the nature of the SARSCoV- 2. Since these residues are in a hyper-variable region and comprise only 6 residues their natural acquisition cannot be underestimated. However, authors suggested an unconventional evolution of SARS-CoV-2.
Transmission and pathogenesis
Exact source of SARS-CoV-2 remains elusive, so does the role of key intermediate host in human transmission. However, since SARS-CoV-2 can recognize ACE2 which is present in variety of animal species including palm civets. In case of SARSCoV, RBM residues were adopted to both human and palm civet ACE2. Such adoption of virus can facilitate replication in more than one host hence paving the way forward to cross specie transmission. However, SARS-CoV-2 do not possess residual changes in RBM region to facilitate binding with ACE2 of civet, likewise based on ACE2 interaction, it was postulated that mice and rats will not serve as an intermediate host for SARS-CoV-2. Therefore, only genetically modified mice and rats may serve as an experimental model. Other animal species including pigs, ferrets, cats, and nonhuman primates possess favorable residues in their ACE2 receptor, increasing the risk of anthropozoonosis and zooanthroponosis transmission of SARSCoV- 2. Hence, domestic pets such as cats and dogs may need attention during an outbreak.
Initially, all the sequenced strains of SARS-CoV-2 were isolated from the patients who visited local food market in Wuhan province except one patient who resided nearby but did not visit local food market directly [8]. It is therefore likely that the source of the viral transmission was an intermediate host in food market, though the genetic origin of the virus seems bats, which served as primary host and humans as terminal host. Hence, its transmission was linked to the seafood market where several infected patients visited or worked [31]. The sea food market sold live animals. Obviously, virus further disseminated via person to person contact mainly through respiratory droplets. However, due to surrounding uncertainty about its transmission, routinely air born precautions are also recommended. It seems as the outbreak progressed, person to person transmission became the main source of the transmission. Transmission has also been reported from individuals with asymptomatic infections or during incubation period [32,33]. The RNA of SARS-CoV-2 has been detected in blood and stool; however, no evidence was provided about the viral particle from these specimens. It has been suggested that the virus is more likely to be detected during convalescent period and can also be transmitted during incubation period [32]. Bats may serve as a reservoir for the coronaviruses in general and particularly for SARS-CoV-2. However, other animals may also serve as an intermediate host between humans and bats [8]. Nevertheless, bats are suggested as an original host for the SARS-CoV-2, yet it is not clear exactly which intermediate host has facilitated the viral infection in humans [8]. Likewise, the exact mechanism of pathogenesis of the disease caused by SARS-CoV-2 is not fully understood. A study reported that the infected patients of SARS-CoV-2 showed higher amounts of IP10, IFNγ, MCP1 and IL1B that leads to T-helper-1 (Th-1) response. However, increase in T-helper-2 (Th-2) cytokines such as IL10 and IL4 had also been observed which play role in suppressing inflammation [34]. Further studies are needed to investigate the pathogenesis of SARS-CoV-2 infection and particularly to characterize the Th-1 and Th-2 response. Recent studies showed that majority of the deaths occurred because of the respiratory failure, However, detailed lung pathology remains to be elucidated [34,35]. The spike protein is an important target for the novel therapy and vaccine [23]. However, complete understanding of spike protein driven immune response for SARS-CoV-2 and its role in the protection remains a significant impediment in this regard. Nevertheless, for other coronaviruses particularly SARS spike proteins are crucial in eliciting immune response and range of neutralizing antibodies along with T-cell response are mediated by S1 epitopic region [36]. It is noteworthy, that most potent antibodies raised against SARS-CoV directed against RBD region remain ineffective against SARS-CoV-2, which is attributed to divergence of RBD region. Hence, identification of novel monoclonal antibodies directed against SARS-CoV-2 remains imperative. Recently identified CR3022 is a novel monoclonal antibody against RBD of novel corona virus. However, monoclonal antibody alone may not suffice a complete therapeutic option since little is known about the SARS-CoV-2 CD4+ T helper cells immune response in respiratory infections [36]. Conclusively, development of novel monoclonal antibodies and molecular mechanism underlying cell mediated immune response remain a crucial aspect for the future research endeavors for the scientific community.
Clinical signs and symptoms
The incubation period for the SARS-CoV-2 is approximately 14 days after exposure and majority of the cases were reported after five days of exposure [8]. Most recent data based on 181 cases of SARS-CoV-2 confirmed incubation period 5 to 6 days and symptoms may appear within 11.5 days [37]. The infected individuals show high grade fever and respiratory symptoms. The most frequent manifestation of the SARS-CoV-2 infection is pneumonia primarily characterized by cough, fever, bilateral infiltrates, and dyspnea [34,18]. Other less common manifestations include shortness of breath, muscle ache, confusion, headache, anosmia, sore throat, chest pain, nausea, vomiting and diarrhea [34]. The variations in the white blood cell (WBC) count such as leukopenia, leukocytosis, and lymphopenia have been observed. The most common condition was lymphopenia [38]. According to the WHO the recovery time in case of mild infection is around two weeks whereas; in case of severe infection it may take three to six weeks [39]. Highest rate of fatalities was observed in comorbid conditions such as immune-compromised, hypertension, obesity, and lung disease.
Laboratory diagnosis
The recommended specimen for the detection of SARSCoV- 2 is nasopharyngeal swab and oropharyngeal swab and when possible sputum, broncholveolar lavage and tracheal aspirate can be used for detection [40]. Other specimens (e.g, stool, urine, and fluids) can also be subjected to analysis. Special airborne precautions should be used while collecting respiratory samples. The Reverse Transcriptase Real Time Polymerase Chain Reaction (RT-PCR) can be used for the rapid detection of SARS-CoV-2 RNA. The virus can be isolated by cell culture using pathogen free human airway epithelial (HAE) cell lines and cytopathic effects can be studied [8]. Genome sequencing can be used for the identification of the SARSCoV- 2. In fact, in near future it must serve as an important tool for the genome-based surveillance to keep a check on the evolutionary development of SARS-CoV-2. In case, initial screening test is negative for the SARS-CoV-2 and suspicious signs and symptoms persist, WHO recommends re-sampling and re-testing from multiple respiratory sites [41]. Serologically anti-SARS-CoV-2 IgG and IgM antibodies can be detected by ELISA Kit [35].
Treatment
There is no specific and proven antiviral treatment of SARSCoV- 2 infection. National Health Commission of China favored combination of anti-HIV drug lopinavir and ritonavir together with a dose of nebulized interferon α for the treatment [42]. Many clinical trials are in progress such as NCT04246242 and NCT04252664 to evaluate the antiviral efficacy and specificity against SARS-CoV-2. The remdesivir is a novel nucleotide analogue that is being used for the treatment in United State. However, further clinical studies on this antiviral drug are currently ongoing [43]. Studies suggest promising activity of remdesivir against SARS-CoV-2 and other coronaviruses like SARS-CoV and MERS-CoV both in vitro and in vivo [42,44]. Although Chloroquine is prescribed in current situation, mechanism of action against SARS-CoV-2 is not fully understood. Current study suggests convalescent plasma therapy in ten patients quite effective in neutralizing viraemia, yet large-scale clinical studies are urgently needed[45].Another study also reported the in vitro effectiveness of FDA approved anti-parasitic drug Ivermectin for the COVID-19 treatment[46].There is no vaccine available for the SARSCoV- 2.
Prevention and control
Prevention must be given the highest priority. To launch the effective prevention strategy first and foremost highest level of vigilance is required for the identification of suspected cases that must be followed by isolation of infected individuals in proper facilities and implementation of infection control measures. Obviously, infection control is the vital component to limit the further transmission of SARS-CoV-2. According to a study in China, an estimated 43% of the 138 patients acquired SARS-CoV-2 in hospital setting [47]. Current situation indicates healthcare workers are at the higher risk if not properly equipped. The suspected individual should be educated to wear mask and seek immediate medical attention. According to the WHO and Center for Disease Control (CDC) guidelines given for the healthcare workers, the standard precautions are avoiding contact, droplet/airborne precautions particularly eyes and face protection and wearing N95 masks [48,49]. The infected individual should be kept in an airborne infection isolation room, in case of unavailability of the facility; the infected individual should wear a mask and be kept in a private room with closed door. The healthcare individuals should wear personal protective equipment while entering the room prior to meeting patient [48]. To reduce the spread of SARS-CoV-2 environmental infection, approved products by environmental protection agency for emerging viral pathogen should be used for disinfection and protection [50]. Other general measures to reduce its transmission include avoiding close contact with infected individuals, respiratory hygiene, avoiding contact with live or dead animals and regular hand washing and use of hand sanitizers. The virus can be inactivated by heating at 56 degree Celsius for 30min or by Ultraviolet (UV) rays and is also sensitive to most disinfectants such as 75% ethanol, diethyl ether, peracetic acid, chloroform, and chlorine [51]. Individuals returning from high risk countries, on arrival should undergo screening for the signs of illness and be monitored (to be kept under quarantine) for at least 14 days. The public health officials must be informed about the suspected cases as soon as possible. Most of all the SARS-CoV-2 poses common challenge to humanity, global coordination is the key to succeed in the race against time. Government needs to take centrally coordinated response towards their societies to limit unnecessary mobility of the individuals. Moreover, it is need of time to outline global policies for food surveillance and handling of wild animals. Initially, the Strategic Preparedness and Response Plan of WHO required total US$675 million out of which US$61.5 million was required on urgent basis from February to April 2020 more funds should be made available to WHO to expedite coordinated efforts against pandemics. Most of all low cost rapid and accurate diagnosis tests at larger scale are the need of the time to effectively control SARSCoV- 2infection.
Conclusion
Conclusively, SARS-CoV-2emerged as an unprecedented challenge. Not much is known about the clinical and epidemiological aspects of infections caused by SARS-CoV-2. Various factors are responsible for the ongoing dissemination of SARS-CoV-2, including human travel, source of infection, transmission mode, viral adaption and unavailability of effective control measures. In recent years, increase in the incidence of coronaviruses such as SARS, MERS and SARSCoV- 2 highlights the need for the effective strategies such as infection control measures, identification of primary and secondary hosts, development of vaccine and new antiviral therapies. Collaborative efforts should be given highest priority to minimize its transmission at global scale. In addition, there is an urgent need to design appropriate diagnostic tests for the identification of the SARS-CoV-2. Furthermore, priority should be given to educate masses and that vigilance and preparedness at national and international level is the key to succeed against future pandemics.
Acknowledgement
None
Disclaimer
None
Conflict of interest
None
Funding source
None
29745
References
- Justyna M, Blicharz-Domańska K (2018) Coronaviruses in Avian Species - Review with Focus on Epidemiology and Diagnosis in Wild Birds. J vet res 249-255.
- Lim YX, Ng YL, Tam JP, Liu DX (2016) Human coronaviruses: a review of virus–host interactions. Diseases 4:26.
- Drosten C, Günther S, Preiser W, van der Werf S, Brodt HR, et al. (2003) Identification ofnovel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med 348:1967-1976.
- Zaki AM, Van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA (2012) Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. New England Journal of Medicine 367:1814-1820.
- Wong G, Shi W, Liu J, Lai ACK, Zhou J, Liu W, et al. (2016) Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol 24:490–502.
- Liu DX, Fung TS, Chong KKL, Shukla A, Hilgenfeld R (2014) Accessory proteins of SARS-CoV and other coronaviruses. Antiviral res 109:97-109.
- Su S, Wong G, Shi W, Liu J, Lai ACK, Zhou J, et al. (2016) Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbial 24:490-502.
- Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, et al. (2020) Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. The Lancet 395:565-574.
- Graham RL, Donaldson EF, Baric RS (2013) A decade after SARS: strategies for controlling emerging coronaviruses. Nature Reviews Microbiology 11:836-848.
- Kelly-Cirino C, Mazzola LT, Chua A, Oxenford CJ, Van Kerkhove MD (2019) An updated roadmap for MERS-CoV research and product development: focus on diagnostics. BMJ global health 4:e001105.
- Widagdo W, Okba NM, Raj VS, Haagmans BL (2017) MERS-coronavirus: From discovery to intervention. One Health 3:11-16.
- World Health Organization Press Conference (2019) The World Health Organization (WHO) Has Officially Named the Disease Caused by the Novel Coronavirus as COVID-19.
- Gorbalenya AE, Baker SC, Baric RS, de Groot R.J, Drosten C, Gulyaeva AA, et al. (2020) Severe acute respiratory syndrome-related coronavirus: The species and its viruses—A statement of the Coronavirus Study Group. bioRxiv.
- Covid-19 Coronavirus Outbreak (2020) Confirmed Cases and Deaths by Country, Territory, or Conveyance.
- Chan JFW, Yuan S, Kok KH, Kai-Wang K, Chu H, Yang J, et al. (2020) A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet 395:514-523.
- Li F (2016) Structure, function and evolution of coronavirus spike proteins. Annual Review of Virology 237-261.
- He Y, Zhou Y, Liu S, Kou Z, Li W, Farzan M, et al. (2004) Receptor binding domain of SARS CoV spike protein induces highly potent neutralizing antibodies implications for developing subunit vaccine. Biochem Biophys Res Commun 324:773-781.
- Li F (2012) Evidence for a common evolutionary origin of corona virus spike protein receptor binding subunits. J Virol 2856 –2858.
- Du L, He Y, Zhou Y, Liu S, Zheng BJ, Jiang S (2009) The spike protein of SARS-CoV — a target for vaccine and therapeutic development. Nature REVIEWS Microbiology 7.
- Xu X, Chen P, Wang J, Feng J, Zhou H, Li X, et al. (2020) Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci. China Life Sci 63:457–460.
- Wan Y, Shang J, Graham R, Baric RS, Lia F (2020) Receptor Recognition by the Novel Coronavirus from Wuhan: An Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. Journal of Virology 94:e00127-20.
- Hoffmann M, Kleine-Weber H, Krüger N, Müller M, Drosten C, Pöhlmann S (2020) The novel coronavirus 2019 (2019-nCoV) uses the SARS-coronavirus receptor 2 ACE2 and the cellular protease TMPRSS2 for entry into target cells.
- Zhou P, Yang XL, Wang ZG, Hu B, Zhang L, Zhang W, et al. (2020) Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin. Nature 579:270-273.
- Morse JS, Lalonde T, Xu S, Liu WR (2020) Learning from the Past: Possible Urgent Prevention and Treatment Options for Severe Acute Respiratory Infections Caused by 2019-nCoV. Chembiochem 21:730-738.
- Yoon V, Fridkis-Hareli M, Munisamy S, Lee J, Anastasiades D, Stevceva L (2010) The GP120 molecule of HIV-1 and its interaction with T cells. Curr Med Chem 17:741-749.
- Rothe C, Schunk M, Sothmann P, Bretzel G, Froeschl G, Wallrauch C, et al. (2020) Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N Engl J Med 382:970-971.
- Kupferschmidt K (2020) Study claiming new coronavirus can be transmitted by people without symptoms was flawed. Science 3.
- Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. (2020) Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet 395:497-506.
- Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, et al. (2020) Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin. BioRxiv.
- Oh HLJ, Gan SK, Bertoletti A, Tan YJ (2012) Understanding the T cell immune response in SARS coronavirus infection. Emerging Microbes and Infections 1:e23.
- Laue SA, Grantz KH, Bi Q, Jones FK, Zheng Q, Meredith HR, et al. (2020) The incubation period of coronavirus disease 2019(COVID-19) from publicaly reported confirmed cases: estimation and application. Ann Intern Med.
- Centers for Disease Control and Prevention (2020) Interim Clinical Guidance for Management of Patients with Confirmed 2019 Novel Coronavirus (2019-nCoV) Infection.
- WHO Director General (2020) Opening remarks at the media briefing on COVID-19.
- Interim Guidelines (2020) Collecting, Handling, and Testing Clinical Specimens from Persons under Investigation (PUIs) for Coronavirus Disease 2019 (COVID-19).
- WHO (2020) Coronavirus disease (COVID-19) technical guidance: Surveillance and case definitions.
- Sheahan TP, Sims AC, Graham RL, Menachery VD, Gralinski LE, Case JB, et al. (2017) Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. SciTransl Med 28:9(396).
- Holshue ML, DeBolt C, Lindquist S, Lofy KH, Wiesman J, Bruce H, et al. (2019) First case of Novel Coronavirus in the United States. N Engl J Med 2020:31.
- Wang M, Cao R, Zhang L, Yang X, Liu J, XuM,et al. (2020) Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res 30:269-271.
- Wan Y, Shang J, Graham R, Baric RS, Li F (2020) Receptor Recognition by the Novel Coronavirus from Wuhan: An Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. Journal of Virology 94:e00127-20.
- Caly L, Druce JD,Catton MG, Jans DA, Wagstaff KM (2020) The FDA-approved Drug Ivermectin inhibits the replication of SARS-CoV-2 in vitro.
- Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, et al. (2020) Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. JAMA 323:1061-1069.
- Centers for Disease Control and Prevention (2020) Interim guidance for persons who may have 2019 Novel Coronavirus (2019-nCoV) to prevent spread in homes and residential communities.
- World Health Organization (2020) Infection prevention and control during health care when novel coronavirus (nCoV) infection is suspected.
- Kampf G, Todt D, Pfaender S, Steinmann E (2020) Persistence of coronaviruses on inanimate surfaces and its inactivation with biocidal agents. J Hosp Inf 104:246e251.
- General Office of National Health Commission (2020) General Office of National Administration of Traditional Chinese Medicine. Diagnostic and treatment protocol for Novel Coronavirus Pneumonia;
- United States Centers for Disease Control and Prevention (2019) Novel Coronavirus Information for Travel.






