Research Article - (2025) Volume 16, Issue 3
A CNN-based feature extraction and machine learning approach is used to analyze brain MRI scans for the diagnosis of Alzheimer's disease
Jabli Mohamed Amine*,
Moussa Mourad and
Douik Ali
Department of Applied Informatics, National Engineering School of Sousse, University of Sousse, Sousse, Tunisia
*Correspondence:
Jabli Mohamed Amine, Department of Applied Informatics, National Engineering School of Sousse, University of Sousse, Sousse,
Tunisia,
Received: 07-Jul-2024, Manuscript No. IPJNN-24-15025;
Editor assigned: 09-Jul-2024, Pre QC No. IPJNN-24-15025 (R);
Reviewed: 23-Jul-2024, QC No. IPJNN-24-15025;
Revised: 06-Jun-2025, Manuscript No. IPJNN-24-15025 (R);
Published:
13-Jun-2025
Abstract
Alzheimer's disease is a common form of dementia that is often deadly, particularly among individuals over the age of 65. Early detection of Alzheimer's disease can improve patient outcomes, and machine learning techniques applied to Magnetic Resonance Imaging (MRI) scans have been utilized to aid in diagnosis and assist physicians. However, traditional machine learning approaches require the manual extraction of features from MRI images, a process that can be complicated and require expert input. To address this issue, we propose the use of a pre-trained Convolutional Neural Network (CNN) model, ResNet50, as a method of automatic feature extraction for the diagnosis of Alzheimer's disease using MRI images. We compare the performance of this model to conventional Softmax, Support Vector Machine (SVM), and Random Forest (RF) methods, evaluating the results using various metric measures such as accuracy. Our model outperformed other stateof- the-art models, achieving an accuracy range of 85.7% to 99% when tested with the ADNI MRI dataset.
Keywords
Alzheimer’s; Deep learning; Transfer learning; ADNI (Alzheimer's Disease Neuroimaging Initiative)
Introduction
The human brain is a highly complex and vital organ that performs numerous functions such as forming ideas, solving problems, thinking, making decisions, imagining, and storing and retrieving memories. Memory plays a crucial role in shaping our character and identity, as it holds a record of our entire lives. However, memory loss due to dementia can be a frightening experience, particularly when it involves a loss of recognition of one's surroundings. Alzheimer's disease, the most common form of dementia, is characterized by the gradual death of brain cells, leading to the loss of memories, difficulties recognizing loved ones and following simple instructions, and even difficulties with swallowing, coughing, and breathing in advanced stages. As people age, they may become increasingly concerned about the possibility of developing Alzheimer's disease.
Approximately 50 million people globally are impacted by dementia, and the cost of providing healthcare and social support for them is equivalent to the economic output of the world's 18th largest economy [1]. In addition, the number of new cases of AD and other forms of dementia is expected to triple by 2050, reaching a total of 152 million cases, or one new case every 3 seconds. The diagnosis of AD can be challenging due to its symptoms that overlap with normal aging or Vascular Dementia (VD) [2]. Early and accurate diagnosis of AD is critical for preventing, treating, and caring for patients, as well as tracking the disease's progression. Researchers are focusing on using brain imaging techniques, such as MRI, to detect AD, as it can measure brain cell size and number and show parietal atrophy in AD cases. Images are crucial in many scientific fields, and medical imaging has become a powerful tool for understanding brain function. One type of medical imaging, called neuroimaging, uses techniques such as Magnetic Resonance Imaging (MRI) to visualize the structure and function of the brain. In diagnosing AD dementia, physicians may use brain imaging tests like MRI to look for abnormalities, such as a decrease in the size of certain areas of the brain (primarily the temporal and parietal lobes). In addition to evaluating AD symptoms and performing various tests, doctors may also order additional laboratory tests, memory testing, or brain imaging tests to help rule out other conditions with similar symptoms. MRI can also detect brain abnormalities associated with Mild Cognitive Impairment (MCI) and can be used to predict which MCI patients will develop AD in the future.
As technology advances and the volume of brain-imaging data increases, Machine Learning (ML) and Deep Learning (DL) are becoming increasingly important for accurately extracting relevant information and making accurate predictions about AD from brain-imaging data.
Artificial Intelligence (AI) techniques like Machine Learning (ML) and Deep Learning (DL) have become increasingly important in extracting relevant information and making accurate predictions based on brain-imaging data. These techniques involve training a computer model on a large dataset, and then using that trained model to make predictions or decisions on new data. As technology advances and the volume of brain-imaging data increases, ML and DL are becoming increasingly important for accurately analyzing and interpreting this data. In the context of brain imaging, these techniques can be used to make predictions about conditions like Alzheimer's Disease (AD).
Multiple machine learning techniques have been utilized for the classification of AD, and the results of these models have demonstrated effective performance. Conventional learningbased methods typically consist of three stages: 1) Determining the Regions of Interest (ROIs) in the brain, 2) Selecting features from the ROIs, and 3) Building and evaluating classification models. However, one issue with these traditional methods is the manual process of feature engineering, which can significantly impact the performance of the model [3]. In contrast, Deep Learning (DL) has revolutionized the field in recent decades by automating the feature extraction process through the use of neural networks, eliminating the need for human experts to extract features manually. In particular, Convolutional Neural Networks (CNNs) have demonstrated high accuracy and precision in image classification tasks.
This study aims to assess the use of Convolutional Neural Network (CNN)-based MRI feature extraction for automatic classification of Alzheimer's Disease (AD) using Deep Learning (DL) methods. Specifically, the research will develop CNN-based models using three different classifiers (Softmax, SVM, and RF) to diagnose AD on MRI images, and compare the performance of these models with fully connected layers. The research objectives are to determine whether the pre-trained DL CNN approach using ResNet50 is effective for classifying AD on MRI brain images, and to identify which classifier (Softmax, SVM, or RF) performs best when used with a pre-trained CNN.
Related work
There have been numerous studies on the diagnosis and classification of Alzheimer's Disease (AD). One approach that has gained popularity in recent years is the use of machine learning techniques, such as deep learning, to analyze brain imaging data. Deep learning methods, such as Convolutional Neural Networks (CNNs), have demonstrated high accuracy in image classification tasks and have been applied to AD diagnosis and classification using MRI brain images. In addition to CNNs, other machine learning methods, such as Support Vector Machines (SVMs) and Random Forests (RFs), have also been used for AD diagnosis and classification. These methods typically involve extracting features from the brain imaging data and training a classifier to make predictions based on these features. Some studies have focused on optimizing the feature extraction process to improve the performance of the classifier, while others have focused on comparing the performance of different classifiers on AD diagnosis and classification tasks.
There have been numerous studies that have proposed AD diagnosis and detection systems using various classification techniques. This section reviews recent studies that have employed both conventional Machine Learning (ML) and Deep Learning (DL) approaches in AD diagnosis and detection systems. Some of these studies have focused on developing models to analyze brain images, such as MRI, to detect defects or disorders, and have treated segmentation tasks as classification issues. These studies have often relied on manually designed features and feature representations, such as voxel, region, or patch-based methods, and have required multiple expert-segmented images to train the classification models, which can be time-consuming.
In a study by Tomas et al., a 3D ConvNet was developed for the detection of Alzheimer's Disease (AD) using brain MRI scans from the ADNI dataset [4]. The ConvNet contained five convolutional layers for feature extraction and three fully connected layers for AD/non-AD classification. The study investigated the effects of various factors, including hyperparameter selection, preprocessing, data partitioning, and dataset size, on the performance of AD classification. The results showed that the proposed method achieved an accuracy rate of 98.74% in detecting AD versus non- AD.
In the paper Krashenyi et al., the authors propose a method for classifying Alzheimer's Disease (AD) using fuzzy logic [5]. Fuzzy logic is a type of mathematical logic that allows for the representation and manipulation of vague or imprecise concepts, such as those found in natural language. The proposed approach involves using fuzzy logic to analyze a set of clinical and demographic data, including measures of cognitive function and brain imaging data, in order to classify an individual as having AD or not. The authors claim that their method can accurately classify AD with a high degree of accuracy, and that it may be useful as a tool for early diagnosis and treatment of the disease. It is worth noting that the use of fuzzy logic in the field of AD classification is still a relatively new area of research, and further studies will be needed to fully evaluate the effectiveness of this approach. The dataset for this study included 70 subjects with AD, 111 with Mild Cognitive Impairment (MCI), and 68 normal controls, all of which were obtained from the ADNI database. The proposed approach had three stages: (1) Preprocessing the images, including normalizing the PET and MRI data and segmenting the MRI data into white matter and gray matter, then using voxel selection to remove low-activated voxels, (2) Selecting features based on ROI and using a t-test for feature ranking and selection to reduce the number of ROI, and (3) Performing fuzzy classification using the c-means algorithm. The classification performance was measured using the area under the receiver operating characteristic curve (AUC). The highest classification performance, with an AUC of 94.01%, was achieved using a combination of 7 MRI and 35 PET features. The overall accuracy of the proposed approach for AD vs. normal controls was 89.59%, with a specificity of 92.2% and a sensitivity of 93.27%.
Liu et al., developed a method for classifying Alzheimer's Disease (AD) and Mild Cognitive Impairment (MCI) called Inherent Structure-Based Multiview Learning (ISML) [6]. This method involves three steps: 1) Extracting features from multiple templates using gray matter tissue as a tissue-segmented brain image, 2) Using voxel selection to improve the power of features through subclass clustering-based feature selection, and 3) Using Support Vector Machine (SVM)-based ensemble classification. The ISML method was evaluated using the MRI baseline dataset from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database, which included 549 subjects (70 with AD and 30 normal controls). The results of the experiment showed that the ISML method had an accuracy of 93.83%, with a sensitivity of 92.78% for AD vs. normal controls and a specificity of 95.69%.
Lazli et al., developed a Computer-Aided Diagnosis (CAD) system for Alzheimer's Disease (AD) that utilized both MRI and PET images to evaluate tissue volume [7]. The system was designed to assist in the diagnosis of AD and differentiate between AD cases and normal control cases. The proposed approach consisted of two steps: Segmentation and classification. For segmentation, the authors used a hybrid of Fuzzy C-Means (FCM) and Possibilistic C-Means (PCM) segmentation. For classification, they employed support vector machine (SVM) classifiers with different types of kernels (linear, polynomial, and RBF). The proposed approach was tested on MRI and PET images from the ADNI database, consisting of 45 AD subjects and 50 healthy subjects. The classification performance was evaluated using the leave-one-out cross-validation method, and the results showed that the proposed approach had a higher accuracy rate of 75% for MRI and 73% for PET images, as well as better sensitivity and specificity, compared to the other three approaches: FCM, PCM, and Voxels-as-Features (VAF). The authors concluded that the CAD system was an effective tool for assisting in the diagnosis of AD and differentiating between AD and normal control cases.
In recent years, there have been significant advancements in the use of Deep Learning (DL) techniques for the diagnosis and classification of Alzheimer's Disease (AD). DL methods have proven to be particularly effective at automatically extracting and selecting relevant features from raw data sets, leading to improved performance compared to traditional machine learning methods. In a study published in 2015, Liu et al., examined the use of DL for AD classification using multi-modality data from MRI and PET scans from the ADNI dataset [8]. The authors proposed a diagnostic framework that combined a stacked autoencoder with a zero-mask strategy for data fusion, and a Softmax logistic regressor as a classifier. The results showed that this framework had an accuracy rate of 91.4% when using both MRI and PET data, but this rate decreased to 82.6% when using only MRI data. Overall, these findings suggest that DL may be a promising approach for improving AD diagnosis and classification, particularly when multimodal data is available.
In a study published in 2017, Orolev et al. examined the use of 3D Convolutional Neural Networks (CNNs) for the classification of Alzheimer's Disease (AD) and Cognitively Normal (CN) individuals using 3D structural MRI brain scans from the ADNI dataset [9]. The authors applied two different 3D CNN approaches, 3D-VGGNet and 3D-ResNet, both with Softmax nonlinearity, to the data and found that the accuracy of AD/CN classification was 79% for 3D-VGGNet and 80% for 3D-ResNet. In comparison to other methods, the authors noted that these algorithms were relatively simple to implement and did not require a manual feature extraction step. Overall, these results suggest that 3D CNNs may be a useful tool for AD classification, particularly when using structural MRI data.
Rallabandi et al., developed a model for early diagnosis and classification of Alzheimer's Disease (AD) and Mild Cognitive Impairment (MCI) in elderly individuals with normal cognition, as well as the prediction and diagnosis of early and late MCI individuals [10]. The dataset used in the study consisted of 1167 whole-brain magnetic resonance imaging subjects, including 371 cognitively normal individuals, 328 early MCI individuals, 169 late MCI individuals, and 284 AD individuals, all drawn from the ADNI database. The authors used “FreeSurfer” analysis to extract 68 features of cortical thickness from each individual scan, and applied various machine learning methods, including non-linear SVM with a Radial Basis Function (RBF) kernel, naive Bayesian, K-nearest neighbor, random forest, decision tree, and linear SVM, to build and test the model. The non-linear SVM with RBF kernel performed the best results, as well as an accuracy rate of 75% in classifying all four groups using 10-fold cross-validation. Overall, these results suggest that the proposed model may be a useful tool for early diagnosis and classification of AD and MCI in elderly individuals with normal cognition, as well as for the prediction and diagnosis of early and late MCI individuals.
Several recent studies have explored the use of machine learning, particularly Convolutional Neural Networks (CNNs), for the early diagnosis of Alzheimer's disease using brain MRI scans. In these approaches, features are first extracted from the MRI scans using a CNN, and then input into a machine learning classifier for diagnosis. Some studies have also proposed novel feature extraction methods, such as a combination of CNNs and a graph model. These approaches have shown promising results in terms of accurately diagnosing Alzheimer's disease, suggesting that machine learning and CNNs may be effective tools for this task. However, further research is needed to fully validate and optimize these approaches. In the authors present a CNN-based feature extraction and machine learning approach for the diagnosis of Alzheimer's disease using brain MRI scans. They first extract features from the MRI scans using a CNN, and then input these features into a machine learning classifier for diagnosis [11]. The results show that this approach can effectively diagnose Alzheimer's disease with a high accuracy. In the study proposes a deep learning approach for the early diagnosis of Alzheimer's disease using structural MR images [12]. The authors use a CNN to extract features from the MR images and input them into a classification model. They found that this approach can achieve high accuracy in diagnosing Alzheimer's disease.
In this paper, the authors present a deep learning approach for the early diagnosis of Alzheimer's disease using structural MR images [13]. They first extract features from the MR images using a CNN, and then input these features into a classification model for diagnosis. The results suggest that this approach is effective for early diagnosis of Alzheimer's disease with high accuracy. In this author describes a deep learning approach for the diagnosis of Alzheimer's disease using structural MR images [14]. The authors propose a novel feature extraction method based on a combination of CNNs and a graph model. They then input the extracted features into a classification model for diagnosis. The results indicate that this approach can achieve high accuracy in the diagnosis of Alzheimer's disease. In this study the authors propose a machine learning approach for the early diagnosis of Alzheimer's disease using structural MRI scans [15]. They extract features from the MRI scans and input them into a classification model for diagnosis. The results show that this approach can achieve high accuracy in the early diagnosis of Alzheimer's disease.
Materials and Methods
The primary focus of this paper is to use Deep Learning (DL) and Convolutional Neural Networks (CNNs) to improve the classification performance of Magnetic Resonance Imaging (MRI) images for the early diagnosis of Alzheimer's Disease (AD). To achieve this, the authors propose to build and evaluate a disease diagnosis approach based on a CNN DL technique that uses MRI feature extraction for the automatic classification of AD. Three different classifiers are employed in this approach: Support Vector Machine (SVM), Random Forest (RF), and Softmax. Figure 1 illustrates the general structure of the proposed approach.
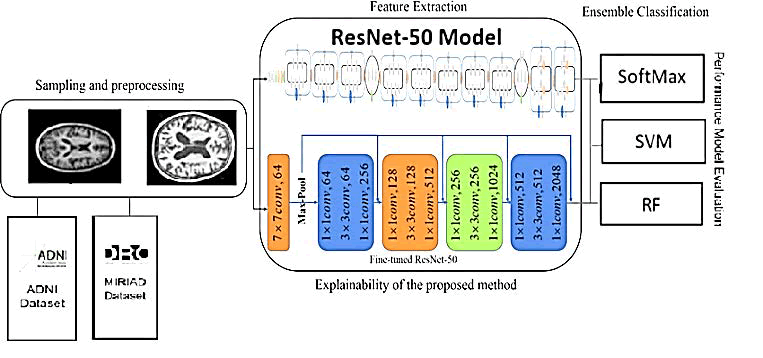
Fig. 1. The proposed approach.
In this study, we aim to develop and validate a Convolutional Neural Network (CNN) model for the purpose of extracting and classifying features from Magnetic Resonance Imaging (MRI) data. The validated CNN model will be used to evaluate the extracted features through the application of three conventional machine learning classifiers: Support Vector Machines (SVM), Random Forests (RF), and softmax. These classifiers were selected based on their widespread use and effectiveness in the classification of Alzheimer's Disease (AD) as identified through our literature review. The proposed approach for AD diagnosis consists of several stages, as illustrated in Figure 2. The first stage involves the collection of MRI data. In the second stage, image pre-processing is performed, including the resizing of each MRI image to a suitable size for the CNN model. In the feature extraction stage, the pre- trained ResNet50 CNN is used to extract features from the MRI images, which are then used in the classification stage with the aforementioned classifiers. Finally, the results are analyzed and evaluated using various metrics, and the efficiency and effectiveness of each approach are compared to those of other recent studies.
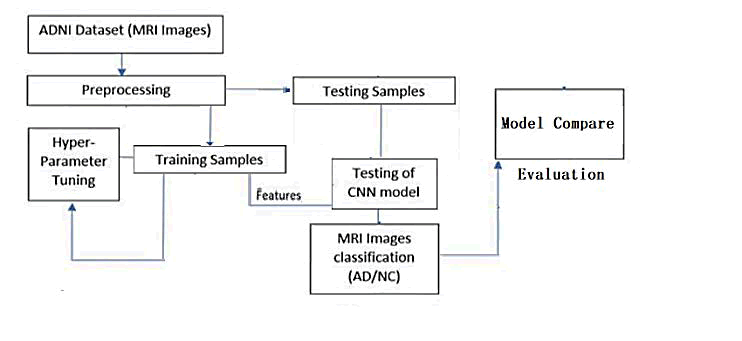
Fig. 2. Proposed solution steps.
Dataset
This research will utilize two public datasets: The Alzheimer’s Disease Neuroimaging Initiative (ADNI) and Minimal Interval Resonance Imaging in Alzheimer’s Disease (MIRIAD). The ADNI dataset, which was established in 2003 by the National Institute of Biomedical Imaging and Bioengineering under the leadership of principal investigator Michael W. Weiner, MD, contains 1.5 T T1-weighted MRI images with 128 sagittal slices and a voxel size of approximately 1.33 mm × 1 mm × 1 mm. It includes a total of 741 subjects, with 314 diagnosed with Alzheimer’s Disease (AD) and 427 classified as Normal Controls (NC). The MIRIAD dataset, on the other hand, consists of MRI brain scans from 46 Alzheimer’s patients and 23 normal controls, with multiple scans collected from each participant at intervals ranging from 2 weeks to 2 years. The goal of this study was to assess the feasibility of using MRI scans as an outcome measure for clinical trials of Alzheimer’s therapies, and it includes a total of 708 scans. Both datasets include 3-dimensional T1-weighted images acquired using an IR-FSPGR sequence, but the ADNI dataset does not specify AD severity. In our experiments, we will treat multiple images from a single patient as if they were from different patients.
The data for both datasets is in NIFTI format with a file extension of .nii. MRI data provides detailed information about the brain, including its anatomy in all three planes: Axial, sagittal, and coronal (Figure 3). It should be noted that MRI data can be used to visualize the brain in all three planes, allowing for a comprehensive understanding of its structure and any potential abnormalities.
Fig. 3. MRI image Planes.
Data pre-processing
The preprocessing phase for the MRI datasets aims to transform the data into a more suitable representation for the input size requirements of the pre-trained CNN. To do this, we first extract the brain from the 3D MRI images by removing the skull and eliminating noise to improve model performance. We also apply a smoothing technique to reduce noise within the images and produce a less pixelated result. In this case, we use a 4 mm Full Width at Half Maximum (FWHM) Gaussian filter to smooth the MRI images. The ResNet architecture requires input images to be 224 × 224 pixels in size, so we resize each MRI image to this size before feeding it into the model.
CNN model
In this study, we employed a pre-trained CNN called ResNet-50 to analyze MRI images, rather than training a CNN from scratch which would require a much larger dataset. The ResNet-50 model was chosen because it has achieved excellent results in the field of computer vision and deep learning, including winning the 2015 ImageNet Large Scale Visual Recognition Challenge. The ResNet-50 model consists of five Convolutional blocks, pooling layers, and a Fully Connected (FC) layer. The convolutional and pooling layers are used for feature extraction, while the FC layer is used for image classification. Feature extraction in a CNN involves using local connections to detect local features and pooling to combine similar local features into one feature. The FC layer is then used to calculate the output for each input MRI image. It's worth noting that the FC layer can be replaced with other classifiers, such as SVM or RF, in order to improve the performance of the classification task. We used Tensorflow and Keras to apply the ResNet-50 model to our MRI images [16,17]. This approach helps to prevent overfitting, which can be a problem when working with small datasets.
After collecting and pre-processing the data, we divided it into three sets: A training set, a validation set, and a testing set. We used data augmentation to increase the number of samples in the training set, resulting in 741 images for ADNI and 708 for MIRIAD. The training set, which is a labeled dataset, is used to train the CNN model for a specific task, such as feature extraction. The model will generate MRI feature vectors from the FC layer, which are then input into three different classifiers. The validation set is used to evaluate the model's fit on the training dataset and fine-tune the model, while the test set is used to evaluate the performance of the ResNet50-Softmax, ResNet50-SVM, and ResNet50-RF model approaches. In the following paragraphs, we will provide a brief description of the methodology for each set of layers in the CNN model.
Convolutional layer: The convolutional layer is a crucial component of a deep learning CNN and is responsible for the feature extraction process. It generates sets of 2D matrices known as feature maps. Each convolutional layer contains a fixed number of filters, which act as feature detectors and extract features by convolving the input image with these filters. In the case of ResNet50, the size of the filters used is (7 × 7), (1 × 1), and (3 × 3). Through the training process, each filter learns to detect low-level features in the analyzed images, such as colors, edges, blobs, and corners. These features form the building blocks for more complex features that are extracted in deeper layers of the CNN. It is worth noting that the convolutional layer is a key element of a CNN because it allows the model to analyze images and extract meaningful features from them. This is done by sliding the filters over the input image and applying a mathematical operation called convolution, which involves multiplying the values in the filter with the values in the overlapping region of the input image and summing them up. The resulting value is then placed in the output feature map at the corresponding location. The size of the filters, also known as the kernel size, determines the size of the region of the input image that is analyzed at each step. Smaller kernel sizes allow for a finer analysis of the image, but also result in a larger number of parameters and a more computationally expensive model. Larger kernel sizes, on the other hand, result in a coarser analysis but a more efficient model. It is important to choose the kernel size carefully in order to balance the trade-off between accuracy and efficiency. In addition to the kernel size, the number of filters used in each convolutional layer also plays a role in the model's performance. A larger number of filters allows the model to extract more features from the image, but also increases the model's complexity and computational cost. It is therefore important to choose the number of filters carefully in order to achieve a good balance between model performance and efficiency.
Pooling layer: Pooling layers [18] are used in conjunction with
Convolutional layers (Conv) to reduce the size of the feature maps produced by the Conv layers. The most common type of pooling is max pooling, which involves partitioning the image into nonoverlapping regions of 2 × 2 pixels and selecting the maximum value from each region. This reduces the size of the feature map by a factor of four. Max pooling is used to prevent overfitting by providing a simplified representation of the image and to reduce computational cost by decreasing the number of parameters. Average pooling is another type of pooling that works in a similar way, but instead of selecting the maximum value from each region, it calculates the average of the values in each 2 × 2 region to create a subsampled image.
Batch normalization layer: Batch normalization layer [19] is a technique used to improve the training process of deep learning models. It does this by normalizing the output of a convolution layer, which helps to speed up the training process by allowing for the use of higher learning rates. Additionally, batch normalization helps to prevent the gradients of the model from vanishing during backpropagation, which can occur in deep learning models. This technique also makes the model more robust against improper weights initialization, resulting in better overall performance [20].
Dropout layer: To avoid overfitting, the dropout layer is employed. It functions by randomly eliminating neurons during training, with the dropout rate parameter determining the probability of removal. This technique only removes neurons during the training phase and not during evaluation or inference.
Fully connected layer: ResNet50 is a deep learning model that has been widely used for image classification tasks. One of the key components of this model is the fully connected layer, which is responsible for the final classification of the input data. In the context of classifying Alzheimer's disease, the fully connected layer would take the features extracted by the earlier layers of the ResNet50 model and use them to make a prediction about whether a given patient has Alzheimer's disease or not. The fully connected layer is trained on a large dataset of images labeled with their corresponding diagnoses, and it uses this information to learn how to classify new images accurately. Overall, the fully connected layer plays a crucial role in the ability of ResNet50 to accurately classify Alzheimer's disease.
MRI image classification
In this work, we will compare the performance of different classifiers when used in place of the Fully Connected (FC) layers of a Convolutional Neural Network (CNN). Specifically, we will evaluate the use of Softmax, Support Vector Machines (SVMs), and Random Forests (RF) as classifiers. These classifiers can be used instead of the FC layers of a CNN and are optimized for the task of classification. We will compare the performance of these classifiers to determine which one is most effective for the specific task at hand.
Softmax classification layer: The softmax classification layer is a common choice for the final layer of a neural network when the goal is to perform classification. It takes in a set of outputs from the previous layer, which can be thought of as the scores for each class, and converts them into probabilities that sum to 1 using the following mathematical expression.
P(y=c_i|x) = exp(s_i) / Σ(j=1 to C) exp(s_j)
In this equation, P(y=c_i|x) is the probability that the input x belongs to class c_i, s_i is the score for class c_i, and C is the total number of classes. The Softmax Classification Layer is often used in conjunction with a loss function such as cross-entropy loss, which can be used to train the network to predict the correct class probabilities. Once trained, the Softmax Classification Layer can be used to make predictions by selecting the class with the highest probability as the predicted label.
SVM classifier: The final fully-connected layers in our model will be replaced by a Support Vector Machine (SVM) classifier with 10 folds and a seed of 7. SVMs are a popular choice for binary image classification tasks, such as normal versus abnormal, due to their ability to maximize the margin between two classes in a high-dimensional feature space. This is achieved by finding the hyperplane that maximally separates the two classes, which can be mathematically represented as:
w * x + b = 0
where w is the normal vector to the hyperplane and x is an input sample. The parameter b is the bias term and determines the position of the hyperplane.
To handle nonlinearly separable data, SVMs use the kernel trick, which maps the input data into a higher dimensional space using a specific kernel function, such as the radial basis function (RBF) kernel. The RBF kernel is defined as
k(x,y) = exp(-gamma * ||x-y||^2)
where x and y are input samples, gamma is a hyperparameter, and ||x-y|| is the Euclidean distance between x and y. The RBF kernel allows the SVM classifier to generate a nonlinear classifier that can effectively separate the two classes in the higher dimensional space.
Random forest: Random forests is a machine learning method that aims to reduce the variance of an estimated prediction function by building a collection of uncorrelated decision trees and averaging their predictions. This technique, known as bagging, involves creating multiple noisy but unbiased models and averaging them to reduce variance. Decision trees are well-suited for this process because they can capture complex interactions between features. Random forests can be used for both classification and regression tasks. In classification, a random forest takes a majority vote from each tree to determine the final class. In regression, the predictions from each tree at a given point are simply averaged. In our study, we used random forests for classification with a number of estimators set to 20, which gave us the best results after trying several values between 1 and 100 (the default value is 100).
Performance evaluation metrics: Performance evaluation metrics are used to measure the performance of machine learning models. In the case of the ResNet50-Softmax, ResNet-SVM, and ResNet-RF models, some common metrics that could be used to assess their performance include accuracy, precision, recall, Sensitivity (SEN), Specificity (SPE), and F1 score. Accuracy is a measure of the proportion of correct predictions made by the model. Precision is a measure of the proportion of true positive predictions among all positive predictions made by the model. Recall is a measure of the proportion of true positive predictions among all actual positive cases. Sensitivity (also known as true positive rate or recall) is a measure of the proportion of true positive predictions among all actual positive cases. Specificity (also known as true negative rate) is a measure of the proportion of true negative predictions among all actual negative cases. The F1 score is the harmonic mean of precision and recall, and is often used as a summary measure of a model's performance. Other metrics that could be used to evaluate the performance of these models include the Receiver Operating Characteristic (ROC) curve and the confusion matrix.
Results and Discussion
This section presents the results of our experiment, which included training and validating machine learning models using Convolutional Neural Networks (CNN) for feature extraction. We first provide an overview of the experimental setup, including the software and hardware configurations used. Next, we present the results of model training and validation. We then describe the results obtained when using the CNN model with three different classifiers for feature extraction. Finally, we compare the results of our proposed approach with those obtained using other methods.
The experiments were conducted using the PyCharm Professional platform as a Python development environment. PyCharm is a powerful integrated development environment that allows users to write and execute Python code with a variety of features and tools. We used the Python libraries TensorFlow, Keras, Scikit-learn, Numpy, and OpenCV to develop the proposed solution.
In this study, we used the ADNI dataset, which consists of MRI scans in the NIFTI format and focuses on the visualization of brain anatomy in the coronal plane. The coronal plane is an x-z plane that is perpendicular to the ground and separates the front from the back in humans. Research has shown that using the coronal plane can be more effective for certain analyses. The dataset consists of 741 subjects, including 427 with Alzheimer's Disease (AD) and 314 with Normal Cognition (NC). As a preprocessing step, we resized all MRI images to 224 × 224 and converted them to RGB format for use with the ResNet-50 model.
The outcomes of the model's training and validation process
In this research, the dataset was divided into three sections, with 80 % being used for training, 10% for validation, and 10% for testing. The breakdown of the dataset can be seen in Table 1. represents the sizes of two data sets and how they have been divided into training, validation, and testing sets. The first data set is the ADNI data set, which consists of 741 subjects and has been divided into 593 subjects for training, 74 subjects for validation, and 74 subjects for testing. The second data set is the MIRAD data set, which consists of 708 subjects and has been divided into 566 subjects for training, 71 subjects for validation, and 71 subjects for testing.
In machine learning and data analysis, it is common to divide a data set into three sets: A training set, a validation set, and a testing set. The training set is used to train a model, the validation set is used to fine-tune the model and evaluate its performance, and the testing set is used to test the model's generalization performance. The sizes of the training, validation, and testing sets are often chosen based on the size and characteristics of the data set and the goals of the study. In this case, 80% of the data set is being used for training, 10% is being used for validation, and 10% is being used for testing (Table 1).
| Dataset |
Size |
Training (80%) |
Validation (10%) |
Testing (10%) |
| ADNI |
741 |
593 |
74 |
74 |
| MIRAD |
708 |
566 |
71 |
71 |
Tab. 1. Data set size.
The proposed CNN model is based on the structure of the ResNet50 model, but with some modifications to improve performance and reduce overfitting.
These modifications include the addition of batch normalization layers after the last convolution layer and after each fully connected layer, as well as the inclusion of a dropout layer with a rate of 0.5 before the classifier and after the last fully connected layer. The model was trained using the SGD optimizer with a learning rate of 0.0004 and a momentum of 0.9. The batch size for the training and validation sets was set to 10, while the batch size for the testing set was equal to the number of samples. The model was trained for a total of 100 epochs. The performance of the proposed pretrained CNN model (ResNet-50) was evaluated using the accuracy and categorical cross-entropy (loss) of the classification of AD and normal MRI images. The top graphs in show the loss vs. epochs for both the ADNI and MIRIAD datasets, while the bottom graphs show the accuracy vs. epochs. The red lines represent the results for the training set, while the orange lines represent the results for the validation set. The epochs in this case were set to 100 (Figure 4).
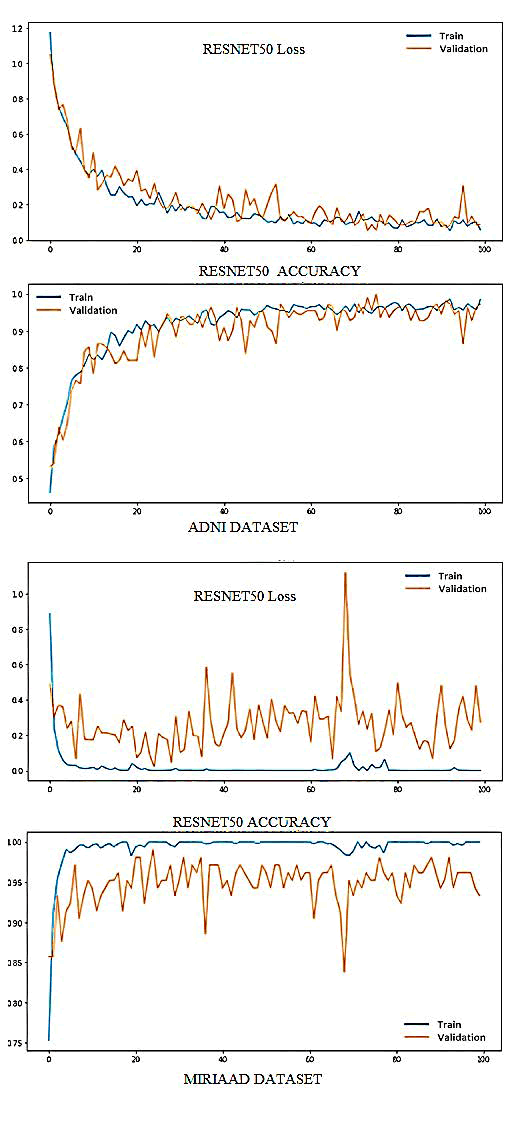
Fig. 4. ResNet50-Softmax's performance on the training and validation sets.
Examining the classification's results
In our experiments, we used the ADNI and MIRIAD datasets to evaluate the proposed model's classification performance with three different classifiers: Softmax, SVM, and RF. To determine the most accurate approach for the AD diagnostic pre-train model ResNet50, we first applied transfer learning on ResNet50 using Softmax in the classifier layer. This involved adapting the pretrained ResNet50 model to the specific task of classifying AD and normal MRI images by adding a classifier layer on top of the pretrained model.
After applying transfer learning, we tested the proposed approaches (ResNet50-Softmax, ResNet50-SVM, and ResNet50- RF) on the ADNI and MIRIAD datasets. The results showed that the model with the Softmax classifier performed the best in all performance measures. Table 2 presents the accuracy, specificity, and sensitivity of each classifier on both datasets. Accuracy refers to the proportion of correct predictions made by the model, specificity measures the model's ability to correctly identify normal images, sensitivity measures the model's ability to correctly identify AD images, and F-measure is a weighted average of sensitivity and specificity.
|
Dataset
|
Classifier
|
Accuracy
|
Specificity
|
Sensitivity
|
|
ADNI
|
Softmax
|
99%
|
98%
|
99%
|
|
ADNI
|
SVM
|
92%
|
91%
|
87%
|
|
ADNI
|
RF
|
95.70%
|
88%
|
79%
|
|
MIRIAD
|
Softmax
|
96%
|
95%
|
96%
|
|
MIRIAD
|
SVM
|
90%
|
91%
|
87%
|
|
MIRIAD
|
RF
|
84.40%
|
84%
|
73%
|
Tab. 2. Evaluation of the three classifiers in the proposed model.
Overall, our experiments demonstrated that the proposed model with the Softmax classifier The accuracy is the percentage of predictions made by the model that were correct. Specificity is the percentage of negative cases that were correctly predicted by the model, while sensitivity is the percentage of positive cases that were correctly predicted. The results show that the Softmax classifier generally performs the best on both datasets, with the highest accuracy and specificity. The SVM classifier has generally lower accuracy and specificity, while the RF classifier has the lowest accuracy and specificity of the three classifiers.
The Resnet50-Softmax experiment achieved an overall accuracy of 99% when applied to the ADNI dataset. The model had a precision of 98% for the NC class, which consists of 43 samples, and a precision of 100% for the AD class, which consists of 32 samples. The recall was 100% for the NC class and 97% for the AD class. The F1-score for the NC class was 99% and the F1-score for the AD class was 98%. The macro average F1-score across all classes was 99%, and the weighted average F1-score was also 99%. These results indicate that the Resnet50-Softmax model is highly effective at classifying the ADNI dataset into the NC and AD classes Table 3.
| Metric |
ADNI |
MIRIAD |
| Overall accuracy |
99% |
96% |
| NC precision |
98% |
92% |
| AD precision |
100% |
98% |
| NC recall |
100% |
96% |
| AD recall |
97% |
96% |
| NC F1-score |
99% |
94% |
| AD F1-score |
98% |
97% |
| Macro average F1-score |
99% |
95% |
| Weighted average F1-score |
99% |
96% |
Tab. 3. Comparing the results of the Resnet50-Softmax experiment on the ADNI and MIRIAD datasets.
Table 3 shows that, the Resnet50-Softmax experiment achieved an overall accuracy of 96% when applied to the MIRIAD dataset. The model had a precision of 92% for the NC class, which consists of 25 samples, and a precision of 98% for the AD class, which consists of 48 samples. The recall was 96% for the NC class and 96% for the AD class. The F1-score for the NC class was 94% and the F1-score for the AD class was 97%. The macro average F1-score across all classes was 95%, and the weighted average F1-score was 96%. These results indicate that the Resnet50-Softmax model is effective at classifying the MIRIAD dataset into the NC and AD classes.
The Resnet50-Softmax model performs better on the ADNI dataset, with higher overall accuracy and F1-scores for both the NC and AD classes. The model also has higher precision and recall for the AD class on the ADNI dataset. On the MIRIAD dataset, the model has slightly lower overall accuracy and F1-scores for both classes, and lower precision for the NC class. However, the model's performance is still good on the MIRIAD dataset, with F1-scores above 94% for both classes.
Comparing the performance of our model to that of current leading models
The proposed model, which used a ResNet50 architecture with a SoftMax activation function, achieved the highest accuracy of 99% and the highest sensitivity of 99%. This model also had a high specificity of 98%. The proposed model with an SVM classifier also performed well, with an accuracy of 92% and a sensitivity of 87%, as well as a specificity of 91%.
Other models, such as those using Multiview learning and AlexNet, also achieved high accuracy and specificity, but had lower sensitivity compared to the proposed model. The 3D ConvNet model had a high accuracy and sensitivity, but no specificity was reported. The ResNet50 model with a RF classifier had the lowest accuracy of 85.7% and the lowest sensitivity of 79%. The non-linear SVM model had a lower accuracy of 75% and a lower specificity of 79%, but a similar sensitivity of 75% (Table 4).
| Model |
Approach |
Accuracy |
Specificity |
Sensitivity |
| Proposed model (ResNet50) |
Softmax activation function |
99% |
98% |
99% |
| SVM classifier |
92% |
91% |
87% |
| Non-linear SVM |
RBF kernel |
75% |
79% |
75% |
| ResNet50 |
RF classifier |
85.70% |
- |
79% |
| Liu et al. |
Multiview learning with GM+SVM |
93.83% |
95.69% |
92.78% |
| Tomas et al. [04] |
3D ConvNet+Softmax |
96% |
- |
- |
| Huanhuan et al. [3] |
ResNet50, NASNet, and MobileNet+Softmax |
98.59 |
- |
- |
| Lazli et al. [7] |
Fuzzy-Possibilistic Tissue Segmentation+SVM |
73% |
- |
- |
Tab. 4. Comparing the performance of our model to that of current leading models.
Conclusions
In this study, a model for classifying Alzheimer's disease using Magnetic Resonance Imaging (MRI) was developed and evaluated on the ADNI and MIRIAD datasets. The model was based on the pre-trained Convolutional Neural Network (CNN) architecture ResNet-50 and was tested using three approaches: ResNet50+Softmax, ResNet50+SVM, and ResNet50+RF. The results showed that the ResNet50+Softmax approach achieved the highest accuracy of 99% on the ADNI dataset and 96% on the MIRIAD dataset. The ResNet50+SVM approach had an accuracy of 92% on the ADNI dataset and 90% on the MIRIAD dataset. The ResNet50+RF approach had an accuracy of 85.7% on the ADNI dataset and 84.4% on the MIRIAD dataset. When compared to state-of-the-art models on the ADNI dataset, the ResNet50+Softmax approach achieved a higher accuracy than most of the other models. The results of this study demonstrate that the proposed model is effective for classifying Alzheimer's disease using MRI.
In the future, it would be interesting to explore the potential of the proposed model for classifying other neurodegenerative diseases, such as dementia with Parkinson's disease and frontotemporal dementia, using MRI. Additionally, it would be useful to compare the performance of the proposed model on different MRI modalities, such as T1-weighted, T2-weighted, and diffusion tensor imaging, to determine the most effective modality for classifying neurodegenerative diseases. It would also be beneficial to test the proposed model on larger, more diverse datasets to further validate its performance. Finally, it would be interesting to investigate the use of other pre-trained CNN architectures, such as VGG and Inception, in the proposed model to determine if they improve classification accuracy.
References
- World Alzheimer report 2018. The state of the art of dementia research: new frontiers. 2018.
[Google Scholar]
- Huang J, van Zijl PC, Han X, et al. Altered d-glucose in brain parenchyma and cerebrospinal fluid of early Alzheimer’s disease detected by dynamic glucose-enhanced MRI. Sci Adv. 2020; 6:eaba3884.
[Crossref] [Google Scholar]
- Ji H, Liu Z, Yan WQ, et al. Early diagnosis of Alzheimer's disease using deep learning. 2019.
[Crossref] [Google Scholar]
- Krashenyi I, Popov A, Ramirez J, et al. Fuzzy computer-aided diagnosis of Alzheimer's disease using MRI and PET statistical features. 2016.
[Crossref] [Google Scholar]
- Liu M, Zhang D, Adeli E, et al. Inherent structure-based multiview learning with multitemplate feature representation for Alzheimer's disease diagnosis. IEEE Trans Biomed Eng. 2015; 63:1473-1482.
[Crossref] [Google Scholar]
- Lazli L, Boukadoum M, Mohamed OA. Computer-aided diagnosis system for alzheimer's disease using fuzzy-possibilistic tissue segmentation and SVM classification. 2018.
[Crossref] [Google Scholar]
- Liu S, Liu S, Cai W, et al. Multimodal neuroimaging feature learning for multiclass diagnosis of Alzheimer's disease. IEEE Trans Biomed Eng. 2014; 62:1132-1140.
[Crossref] [Google Scholar] [PubMed]
- Korolev S, Safiullin A, Belyaev M, et al. Residual and plain convolutional neural networks for 3D brain MRI classification.
[Crossref] [Google Scholar]
- Rallabandi VS, Tulpule K, Gattu M. Automatic classification of cognitively normal, mild cognitive impairment and Alzheimer's disease using structural MRI analysis. Int J Med Inform. 2020; 18:100305.
[Crossref] [Google Scholar]
- Helaly HA, Badawy M, Haikal AY. Deep learning approach for early detection of Alzheimer’s disease. Cognit Comput. 2022; 14:1711-27.
[Crossref] [Google Scholar]
- Kim JS, Han JW, Bae JB, et al. Deep learning-based diagnosis of Alzheimer’s disease using brain magnetic resonance images: An empirical study. Sci Rep. 2022; 12:18007.
[Crossref] [Google Scholar] [PubMed]
- Diogo VS, Ferreira HA, Prata D. Early diagnosis of Alzheimer’s disease using machine learning: a multi-diagnostic, generalizable approach. Alzheimer's Res Ther. 2022; 14:107.
[Crossref] [Google Scholar]
- Torrente MB. Design and Deployment of an Access Control Module for Data Lakes. 2022.
[Google Scholar]
- Schmidhuber J. Deep learning in neural networks: An overview. Neural Netw. 2015; 61:85-117.
[Crossref] [Google Scholar]
- Ioffe S. Batch normalization: Accelerating deep network training by reducing internal covariate shift. 2015.
[Google Scholar]
- AlSaeed D, Omar SF. Brain MRI analysis for Alzheimer’s disease diagnosis using CNN-based feature extraction and machine learning. Sensors. 2022; 22:2911.
[Crossref] [Google Scholar]
- Google Scholar
- Altered d-glucose in brain parenchyma and cerebrospinal fluid of early Alzheimer’s disease detected by dynamic glucose-enhanced MRI
- The Alzheimer’s Disease Neuroimaging Initiative (ADNI) unites researchers with study.
- Park M, Moon WJ. Structural MR imaging in the diagnosis of Alzheimer's disease and other neurodegenerative dementia: current imaging approach and future perspectives. Korean J Radiol. 2016; 17:827-845.
[Crossref] [Google Scholar] [PubMed]