Keywords
Antibiotic susceptibility; Bacteria; Penicillin; Pathogen
Introduction
The accidental discovery of the first commercial antibiotic “Penicillin”, by Alexander Fleming in 1928, has saved millions of lives worldwide. Penicillin has been called the miracle drug and was considered a boon to both the scientific community and the general population alike. Since then, several antibiotics have been discovered and as an effect, diseases and infections once considered life threatening and fatal can now be treated easily and cured. The versatile nature of these antibiotics has also lead to its wide-spread usage not just in the healthcare industry but also in food and animal industries. However, they have been exploited by wide variety of industries for too long and far too frequently. Day by day, the over and nonspecific administration of antibiotics is leading to accelerated development of antibiotic resistances among bacterial pathogens [1,2]. Those bacteria that have developed resistance genes (either intrinsic or acquired) can also transfer their resistance to other bacteria through horizontal gene transfer leading to spread of resistance from one organism to the other [3]. In simple terms, antibiotic resistance occurs when the microorganisms develop means to defend against the negative effects of specific antibiotics, hence preventing the antibiotics from effectively killing them. This typically occurs as a process of natural selection, when bacterial colonies are exposed to sub-optimal concentration of antibiotics or as a result of unnecessary/inappropriate prolonged use of antibiotics, both of which exert selective pressure on the bacterial colonies to specifically allow resistant bacteria to emerge. As a result, antibiotics that are designed to kill the bacteria are also the very reason to render them resistant.
In the recent years we have seen a drastic increase in the antibiotic resistance among bacterial pathogens and this is considered as one of the biggest threats to global health in the current era [4]. Antibiotic resistance affects 2 million people per year in the US alone and leads to at least 23,000 deaths [5]. Worldwide, it leads to 700,000 deaths each year and experts predict the number could grow to 10 million deaths annually by 2050 [6], unless stringent actions are taken to curb misuse and overuse of antibiotics. Data also suggests that, although antibiotic resistant bacterial infections can occur in the community, most deaths due to resistance are seen among inpatient healthcare settings such as hospitals and nursing homes [5].
In a hospital setting, when a patient shows signs of infection such as sepsis, they are promptly placed on empiric broad spectrum antibiotics. Current literature indicates that about 51% of the patients in general wards and 82% of the patients in Intensive Care Units (ICUs) are already on antibiotics within 4 hours prior to collection of blood samples for culture [7]. After the collection of blood samples, the patients’ samples are transferred to the clinical microbiology lab to be tested for the presence of microorganisms by incubating in a blood culture instrument that monitors growth. If the sample is flagged positive, further tests will be conducted to identify specific bacteria and their antibiotic susceptibility profile. Common clinical protocol for positive patient samples involve gram stain of the culture sample for rapid, general identification of the organism, followed by streaking them onto agar plates to obtain pure isolates of bacteria which can take anywhere from 24 hours to several days depending on the growth rate of specific bacteria. These isolated bacterial colonies are then tested for their identification (ID) and antibiotic susceptibility profiles against a panel of different antibiotics at varying (serial) concentrations which can take an additional 1-3 days for complete reporting of results.
During this wait time, patients continue to receive broad spectrum antibiotics, increasing the chances for development of antibiotic resistance. This can be averted if specific targeted antibiotics can be administered to the patients in the early stages of infection, thereby reducing the use of broad spectrum antibiotics. Every hour of delay in administrating the targeted antibiotics to septic shock patients, decreases their chances of survival by 7.6% [8]. Hence, obtaining rapid Antibiotic Susceptibility Testing (AST) results to determine the Minimum inhibitory concentration (MIC) values are of high priority in any clinical setting.
MIC is defined as the minimum concentration of the antibiotic which prevents visible growth of a microorganism in a agar or broth dilution susceptibility test [9]. These MIC values in combination with bacterial ID are required to obtain antibiotic susceptibility interpretations and breakpoints. A breakpoint is defined as the selected concentration of the antibiotic which provides interpretation of whether the species of the bacteria is susceptible or resistant to the antibiotic [10]. Bacteria are considered susceptible if the MIC value for the antibioticbacterial pair is lower than the breakpoint and are considered resistant if the MIC value is above the breakpoint, while for the MIC values in between, it is declared as intermediate susceptible. These breakpoints for each bacteria-antibiotic pair are predetermined in accordance with the Clinical Laboratory Standards Institute (CLSI) in USA and European Committee on Antibiotic Susceptibility Testing (EUCAST) in Europe. These numbers provide valuable information to physicians to determine the appropriate targeted antibiotic to be administered to the patient. It is important to note here that, just having either bacterial identification or AST alone, will not yield clinically significant reports for patient treatment. The combined results from bacterial identification and AST are imperative to meaningfully determine the right antibiotic choice for that particular pathogen [11]. The current AST methods practiced in the clinical microbiology labs are accurate, but are either labor intensive or time consuming, leading to long wait times to obtain AST results.
As represented in Figure 1, when samples are received in clinical microbiology lab, depending on the type of sample they are either cultured in broth media or inoculated onto agar plates directly. For example, in case of blood sample from patients suspected of sepsis, (where the concentration of the bacteria is <103-4 CFU/ml of pathogens, they are cultured in broth media initially to detect the presence of pathogens and then subsequently plated onto agar plates for isolation, while in the case of urine samples from patients suspected of UTIs (where the concentration of the bacteria is >103-4 CFU/ml), they can directly be cultured onto agar plates to isolate the pathogens (bypassing the broth culture), to be used for downstream pathogen identification and AST determination.
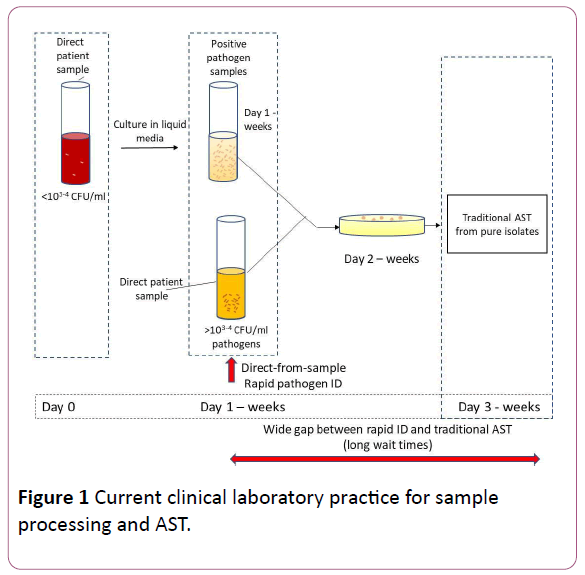
Figure 1: Current clinical laboratory practice for sample processing and AST.
With recent advancements, a number of products have been developed which can provide direct-from-sample pathogen identification (when the concentration of the pathogens in the sample is 103-4 CFU/ml or higher), bypassing the need for isolating the colonies. Examples include the BioFire FilmArray®(Biomerieux), the ePLEX™ Blood Culture ID Panel (Genmark Diagnostics), the Verigene Bloodstream Infection Panel (Luminex Corp), etc. In addition to pathogen identification, many of these also provide information regarding presence or absence of a select set of genes known to cause antibiotic resistance. For instance, the BioFire provides information on 3 genes: mecA, vanA/B and Klebsiella pneumoniae carbapenemase (KPC). Many clinical microbiology labs possess one or more of these instruments. Knowing the presence or absence of such resistance genes guides physicians to some extent to select relatively narrow spectrum antibiotics, but the situation is still far from ideal because finalized and complete susceptibility results are needed to enable definitive antibiotic de-escalation. Hence, despite the ability to perform rapid (same day) identification of bacteria, clinicians will still have to wait a day or more (sometimes >1week) to obtain the final piece of the puzzle (phenotypic AST results, i.e. MIC values) to begin targeted therapy (Table 1).
| System |
Methodology |
Time taken for AST |
Direct from sample |
| Manual systems |
| Broth microdilution |
Media (in ml) containing different antibiotics tested against pathogen of interest |
24 hours |
No |
| Broth microdilution |
Media (in µl) containing different antibiotics tested against pathogen of interest |
24 hours |
No |
| Agar dilution |
Antibiotic incorporated into agar plates and bacteria inoculated on surface |
16–20 hours |
No |
| Disk Diffusion |
Antibiotic impregnated filter discs placed on agar surface pre-inoculated with pathogen |
16–24 hours |
No |
| Etest |
Plastic strip impregnated with gradually decreasing concentrations of antibiotic placed on agar surface pre-inoculated with pathogen |
24 hours |
No |
| Automated systems |
| Vitek system |
Measures light attenuation by optical scanner for growth/no growth detection in micro-wells with different antibiotics |
6–36 hours |
No |
| BD Phoenix system |
Uses redox indicator for detection of growth in micro-well panels containing various antibiotics at different concentrations |
4–16 hours |
No |
| Sensititre |
Fluorescence technology used to monitor activity of enzymes produced by test organism emitting fluorescence |
18–24 hours |
No |
| Microscan Walkaway |
Colorimetric readings using photosensors for optical detection of bacteria |
4.5–7 hours |
No |
| Emerging Technologies |
| BacterioScan FLLS |
Uses laser light source with scattered intensity measurements for accurate OD readings in the presence of antibiotics |
6–18 hours |
Yes |
| Smarticles Technology |
Detects increase in luciferase activity due to bacterial growth from plasmids containing DNA probes inside phages |
<4 hours |
Yes |
| Accelerate Pheno system |
Dark-field microscope used to image cells to record growth vs. no growth vs. cell lysis |
~7 hours |
Yes |
| LifeScale system |
Changes to cantilever vibration changes measured by sensor and correlated to biomass can determine MIC |
~ 3 – 4 hours or longer |
No |
| Future Technologies |
| AFM Cantilever |
Measures changes in amplitude of cantilever fluctuations based on immobilized bacteria on the surface of cantilever |
< 1 hour |
No |
| MAC system |
Bacteria immobilized in agarose and monitored using real-time time-lapse microscopy |
4-10 hours |
Unsure |
| SERS-AST |
Uses silver nanoparticles in nano-channels to identify SERS spectroscopic patterns to determine MIC |
2 hours |
No |
| fASTest |
Uses microfluidic channels into which bacterial cells are trapped and monitored for growth with microscopic imaging |
< 1 hour |
Yes (urine) |
| Isothermal microcalorimetry (IMC) |
Measures heat flowrate of bacterial samples in the presence or absence of antibiotics |
~ 24 hours |
Yes |
| m-EIS system |
Uses microfluidic impedance measurements to determine capacitance changes in presence of different concentrations of antibiotics |
< 4 hours |
Yes |
Table 1: Summary of the current, emerging and future AST technologies.
Rapid AST results (direct-from sample or otherwise) may help reduce the overuse of broad-spectrum antibiotics and thereby curtail the development of drug-resistance. By enabling earlier institution of targeted therapy, it carries the potential to improve patient outcomes in terms of mortality, adverse effects, duration of hospitalization and hence health care costs. There are a few rapid identification systems in the market currently, which can directly use positive culture samples to determine bacterial identification, but none exist for rapid AST. This has not gone unnoticed among the scientific community and several researchers are working on developing rapid AST diagnostics to improve clinical outcomes and reduce antibiotic resistance among microorganisms. A few of the current, emerging and future technologies under development in this field have been discussed in detail in this article.
Current technologies
Most current technologies in the field of Atypically use bacterial growth and/or metabolism to determine the effect of antibiotics. Some of the common methods include agar dilution, disk diffusion, Etest® etc., which typically allow bacteria to grow to confluence and interpret growth/no growth to determine the susceptibility. One such common method that has been used for decades is the broth dilution method.
Manual systems in clinical microbiology
Broth macro-dilution method: This method uses multiple tubes, each containing doubling concentrations of antibiotic being tested. The volumes used per tube are typically 1 ml or higher and hence is considered a macro-dilution method [12]. The bacteria of interest are isolated to obtain single colonies on an agar plate, suspended in media, diluted appropriately and added to each tube to obtain a final concentration of ~5×105 CFU/ml, as per the CLSI recommendation. The tubes, now containing media, bacteria and antibiotics at varying concentrations, along with one positive control tube (with bacteria and media but no antibiotic) are incubated to allow for optimum growth of bacteria for a period of 24 hours or longer. Following this, they are observed for growth either visually or by optical methods. The lowest concentration of each antibiotic at which no visible bacterial growth is observed in the tube is determined as the MIC. This method is easy to interpret and accurate in identifying the MIC for a given bacteria-antibiotic combination but is labor and material intensive and also time consuming due to the difficulty in running multiple samples simultaneously to test a wide panel of pathogens.
Broth microdilution method: This utilizes the same principle as the broth macro-dilution method but is run at microliter volumes. The typical volumes in these wells are in microliter scale (~100 μl) [12]. Here, the samples are dispensed into microtiter well plates, typically containing 96 wells (12×8) as an array with each row containing a given antibiotic at doubling concentrations and every row having a different antibiotic of interest. Due to its miniaturization and small volumes, multiple drugs and/or bacteria can be tested simultaneously on a single micro well plate. The wells however still need to be incubated for a minimum of 12-24 hour period, at the end of which they are read for turbidity either visually or by using automated readers. A commonly used automated system that employs the micro-dilution method is the Micro-naut AST system. Here, dried antibiotics at different concentrations are placed in the microtiter wells, which get dissolved in the bacterial suspension when it is added. These plates are incubated for period of 6-24 hours and read using a photometer [13]. The results are interpreted and reported as breakpoints, MICs or combination of both. Such a method can handle multiple samples or test multiple antibiotics simultaneously using small volumes. The panels used can be customized, pre-made and hence enable ready utilization in labs without extensive capability to run other AST methodologies. However, the time taken to obtain the results is similar in comparison to microdilution method.
Agar dilution method: This is a well-established method where the antibiotic to be tested against a given bacteria is incorporated directly into the agar medium. The bacteria are then inoculated on the surface of the agar plate as 104 CFU spots, typically 5-8 mm in diameter. These plates are allowed to incubate and grow for a period of 16-20 hours or longer. The lowest concentration of antibiotic plate on which no bacterial growth is observed is considered as the MIC. Although only one concentration of antibiotic can be tested per agar plate in this method, multiple organisms can be tested on a single plate using inoculum replicators, which can transfer 32-36 inocula per plate [12]. However, this method still needs manual inspection to determine MIC values and hence can sometimes be misinterpreted if the inhibition zone is not discernable. There are also certain antimicrobials (ex. Colistin/Polymyxin, Sulfa antibiotics) which do not lend themselves well to agar plate testing.
Disk diffusion method: In this method, the bacterial sample isolated from the patient is spread onto a fresh agar plate using swabs. Multiple pre-determined concentrations of antibiotic impregnated filter discs are then placed on the surface of the inoculated agar with good spacing and are incubated at 37°C for a period of 16-24 hours. The diameter of the zone of clearance around the disc is measured and compared to the CLSI reference table to determine if the organism is susceptible, intermediate or resistant against the antibiotic agents tested [14]. This method can test multiple drugs or concentrations on a single agar plate but only yields qualitative results since it doesn’t determine the MIC values which is of high clinical significance for effective patient treatment.
Etest®: This is one of the commonly used gradient diffusion methods developed by bioMérieux, where a plastic strip impregnated with gradually decreasing concentrations of a given antibiotic is placed on the surface of an agar plate preinoculated with bacteria to be tested. The strip has an interpretive scale on the other side, which aids in reading the zone of inhibition. The plates are incubated for ~24 hours, at the end of which the inhibition zone is identified and the corresponding MIC value is determined. The MIC is interpreted as a point on the scale of the strip where the inhibition zone intersects the strip [15]. This method can be used to test the effect of multiple antibiotics per plate, when placed at sufficient distance from each other to prevent inhibition zone overlaps. However, only one organism can be tested per plate and time taken to yield results is comparable to other agar diffusion or dilution methods. The method requires visual interpretation of MIC which can at times be misleading if the zones of inhibitions overlap.
Due to the uncertainties in result interpretation, labor intensiveness and long times taken to obtain results, there has been an increasing trend towards use of automated systems to determine antibiotic susceptibility profiles, which are more reliable and easy to use. There are currently several such systems available in the market targeted towards reducing the sample processing times and effort, automated result interpretations and easy integration with the laboratory information systems (LIS). A few of the commonly used automated systems in the clinical microbiology labs are discussed in detail in the following section.
Automated systems in clinical microbiology
VITEK® system: This system was originally developed in the 1970s by bioMérieux for determining bacterial identification (ID) and AST profiles simultaneously from isolated patient samples. The current systems in the market are VITEK® 2 compact and VITEK® 2 systems both of which utilize broth microdilution technique. The system uses “AST cards” which contain microwells with fluidic connections to automatically fill the samples into multiple wells simultaneously. Each card contains 64 microwells that are loaded with dehydrated culture media and antibiotics at different concentrations. The card also includes one well which only contains dehydrated culture media without any antibiotic, to be used as a positive control well. This is a fully automated system which uses attenuation of light measured by an optical scanner for growth or no growth detection [16]. Since the system uses optical method for detection, it is essential that the samples placed into cards should be pure microbial isolates. A representative isolate is first obtained from colonies in the inoculated agar plates, then suspended in saline solution and adjusted to obtain 108 CFU/ml of microbial concentration. This vial of bacterial suspension is coupled with an AST card, scanned and placed into the VITEK® system. The suspension is diluted automatically to obtain 5×105 CFU/ml, filled into the VITEK® cards, sealed and incubated within the instrument. The instrument periodically monitors for growth in each well over a period of 18-24 hours for bacteria and 36 hours for yeast [17]. The MIC values are determined based on optical observation of growth or no growth in individual wells and a MIC table for different antibiotics along with its interpretation (If bacterial ID is known) is generated and reported by the system. The VITEK® system can handle 120 cards with their XL system and 15/30/60 cards with the VITEK® Compact system.
This system is one of the commonly used automated systems in clinical microbiology labs for simultaneous determination of bacterial ID and AST due to its ease of use and reduced manual labor. It has been easily integrated into the clinical workflow and has decreased the turnaround time in clinical laboratories. However, since the system uses optical detection method, a positive culture sample needs to be plated onto the agar plates, incubated for 1-5 days to obtain pure colonies before they can be utilized by the VITEK® system. Thus, these steps add to the total time taken to obtain AST results once the positive sample has been identified, limiting its time savings. Additionally, there are some organisms that do not key out correctly or yeild unreliable MIC reports (ex.Pseudomonas).
BD Phoenix™ automated identification and susceptibility testing system: This is another automated microdilution-based system employed in clinical microbiology labs and approved by the Food and Drug Administration (FDA) for AST determination. The assay uses redox indicator for detection of growth of organisms in the micro-well panels [18]. Each micro-well contains an antibiotic at a particular concentration which is rehydrated with addition of bacterial suspension. These panels are incubated over a time period and scanned for microbial growth using chromogenic or fluorogenic substrates. Each panel contains an ID and an AST section, each with multiple microwells. The AST section of the panel consists of 84 wells including 1 positive control well. The BD phoenix system is capable of reading 99 AST panels and has a dedicated expert software system which has a series of rules used to report MIC value for given antibiotic along with susceptible, resistance or intermediate interpretation. This system takes between 4-16 hours to obtain MIC values depending on the type of microorganism. Due to its automation, the system eliminates drawbacks of the manual systems and is easy to operate. This system however still requires pure isolated culture of bacteria for AST determination and interpretation, which is a timeconsuming factor.
Sensititre™: This is a commercially available product by Thermo Fisher Scientific based on microdilution method similar to VITEK® and the Phoenix™ systems. The actual detection of growth/no growth of bacteria is customizable for the results to be read manually, semi-automated or fully automated. For manual interpretation, the wells are observed for visual turbidity, while for automated detection, fluorescence technology is used to monitor activity of specific enzymes produced by the organism over the incubation time. The enzymes produced cleave the bond between the fluorophore and the quencher substrate, releasing the fluorophore to emit fluorescence [19]. The amount of fluorescence is directly related to the growth of the organism and is used to report the MIC interpretations. The fully automated system is capable of handling multiple samples simultaneously and hence has a high sample throughput taking between 18-24 hours to obtain results. The system allows for customizable plates with multiple antimicrobials on a single format reducing the need for offline testing due to inadequate result.
Micro-scan walk away®: This is an automated system by Beckman-Coulter for bacterial ID and AST based on broth microdilution method. This system is available in 40 and 96 panel modules, for medium and large-scale operations. It utilizes colorimetric readings based on usage of photosensors and color wheel/lamp assembly for optical detection of bacteria in the wells [20]. Similar to other products, these also have dried antibiotics along with media, which is rehydrated by inoculation of bacterial suspension and incubated to determine growth or no growth in individual wells. The threshold concentration for bacterial detection in this system is 2×107 CFU/ml [20]. Thus, AST profiles for fast-growing organisms can be determined in 4.5 to 7 hours, while the same can take up to 18 hours for slowgrowing organisms. The system is able to deliver accurate information for both microbial identification and AST determination. It also allows for simultaneous processing of conventional as well as specialty panels on a single platform.
The automated systems are now routinely used in the clinical laboratories due to the associated ease of use and smooth workflow. These systems eliminate the uncertainty from result interpretation relative to manual methods and reduce sample handling times. However, since they require pure bacterial isolates and use optical methods for interpretation of results, they typically take about the same time as manual methods to obtain antibiotic susceptibility profiles. Hence, there are newer methods being developed by researchers to reduce the dependency on acquiring pure isolates that use innovative techniques to obtain antibiotic profiles aimed at drastically cutting down the time taken to determine the MIC values.
Emerging technologies
These are technologies that are currently in the process of being commercialized to be used in clinical laboratories. These methods are mainly focused on reducing the sample processing times to obtain faster AST results. Some of these methodologies the advantages and their caveats are discussed in this review below.
BacterioScan™ FLLS: This is one of the emerging technologies for early determination of AST and uses forward laser light scatter technology (FLLS). This system uses a laser light source to measure the concentration of particles in the liquid samples (optical density-OD) as well as the scattered intensity in a direction near to the laser beam. Due to the use of FLLS, the OD can be measured accurately up to 2 orders of magnitude lower than traditional methods leading to low threshold concentrations of ~ 10,000 CFU/ml [21] in comparison to current methods. The system can be used to run up to 16 samples simultaneously, with measurements done automatically every 3 minutes for accurate density change measurements. This was initially developed for urinalysis and now has been adapted to be used for antibiotic susceptibility testing. Using this technology, the system can obtain results in about 6 hours for fast growing microorganisms and can take up to 18 hours for slower growing organisms. This method has been compared to VITEK® and MicroScan® systems and has high rate of agreement on the result interpretations. The system is accurate and has potential to replace the existing microdilution systems and is not limited by the use of probes or markers. The main disadvantage of the BacterioScan™ FLLS is that it cannot distinguish between live and dead bacteria in the samples, and hence good statistical analysis and mathematical modeling will be necessary to eliminate background and baseline disturbances from the readings.
Smarticles™ Technology: This is a rapid molecular diagnostic based method being developed by Roche, which is used to determine antibiotic susceptibility. The technique uses DNA probes inside non-replicating bacteriophages that can specifically bind to particular bacterial genus combined with synthetically designed plasmids. These plasmids contain luciferase gene which gets activated on contact with drugresistant bacteria [22]. The increase in luciferase activity is directly related to the increase in the bacterial numbers in the sample. When this method is used against a panel of different antibiotic concentrations, it can lead to MIC determination. The main advantage of this method is that it can be used to determine antibiotic susceptibility panels directly from positive blood culture samples bypassing the need for bacterial isolation. The method claims to obtain results in less than 4 hours using this Smarticles technology, thereby giving the healthcare providers the knowledge to determine the best antibiotic course to be given to patients early.
Accelerate Pheno™ system: This is a fully automated AST system developed by accelerate diagnostics and is the first system to be approved by FDA for AST determination directly from patient samples. It involves an automated sample preparation step, where the sample is passed through a gel electro-filter, which separates out the used blood cells, culture media and other debris while retaining the bacterial and yeast cells [23]. The purified microbial cells are released into culture media, introduced to multichannel cassettes where they are immobilized. Each of these multi-channels contains different antibiotics at different concentrations. At the point of immobilization, a dark-field microscope is used to image the cells every 10 minutes to record the growth of the cells vs no growth or cell lysis over time. These time-lapse images are then analyzed using their custom software and the MIC values are determined. This automated system can perform both ID and AST within about 7 hours [11]. This system is rapid and can directly use positive blood cultures eliminating the need for pure isolates, thereby saving significant time. However, this automated system in its current form can only process one sample at a time and hence may slow down the clinical work flow. The technician may need to wait for the results on the first sample to be reported before the next sample is input, counteracting any savings in time obtained by rapid AST.
LifeScale® system: The lifeScale® system being developed by Affinity Biosensors uses micro-cantilevers over which microbial cell suspension is passed through. As the cells pass through the resonator, the frequency of the cantilever vibration changes, which is measured by the sensor and correlated to the biomass of the bacteria. By measuring the individual biomass and the count of number of microbes per sample volume (concentration), one can accurately determine the antibiotic susceptibility profile of these microbes [24]. In samples without any antibiotic, the total biomass will increase over time, while in samples with antibiotic concentrations at or above the critical concentrations, there will be an observed loss in total biomass and mean mass of microbes. Thus, by comparing these values at different time points, the system will be able to determine the resistance/susceptibility profiles for a given bacteria-antibiotic combination. This method can be used to obtain results directly from positive blood culture samples and direct urine samples where concentrations of bacteria are above 104 CFU/ml. The results obtained are directly dependent on the doubling time of the bacteria and hence the time taken to obtain the results can vary. Fast growing bacteria with 20-30 minute doubling times take ~ 3-4 hours to achieve AST results, while slower growing bacteria will take longer.
Future technologies
Scientific advances are crucial in any field and more so in a healthcare setting where constant innovation can not only make life easier for its end users but also provide better compensation, lead to better patient outcomes and improved quality of life. Thus, there is a constant need to upgrade the existing technologies and develop innovative new methodologies for better, safer and faster diagnosis of diseases. Below we discuss a few innovative approaches still in development for rapid AST aimed at faster detection times and reduced sample processing for effortless integration into a clinical lab setting.
Atomic force microscopy (AFM) cantilever: The AFM cantilever method characterizes the real-time physical activity of the bacteria utilizing low frequency fluctuations of the cantilever. The bacteria to be tested are immobilized on the surface of the cantilever and their movement causes an increase in the amplitude of the cantilever fluctuations which is sensed by the sensing chamber. As the bacteria are exposed to the antibiotics to which they are sensitive, their activity decreases, leading to a corresponding decrease in the cantilever fluctuations [25]. This can be used to identify specific concentrations of antibiotics at or above which the bacteria become susceptible. The authors were able to determine this pattern by exposure of antibiotics to the bacteria within 15 minutes. The resistant bacteria showed either an increase in the fluctuations of cantilever or initial drop in activity due to metabolic shock followed by return to normal cellular activity, while susceptible strains showed a decrease in their activity. This approach can determine AST profiles of fast growing bacteria within an hour and is one of the rapid methods mentioned in this review. This process is one of the most rapid methods mentioned in this review and can be used to develop quantitative anti-biograms. However, this promising methodology may need pure isolates of bacteria or sample preprocessing as the presence of other non-bacterial cells in direct patient samples may affect bacterial immobilization or cantilever fluctuations.
Microfluidic agarose channel (MAC) system: This system uses immobilized bacteria in the agarose media to determine AST. The bacteria are mixed with liquid agarose and then injected into the microfluidic channel, which immobilizes them inside the channel. A capillary valve is used to introduce the media and antibiotics into the agarose which slowly diffuses into the agarose matrix. The section of this matrix is monitored in realtime using microscopy. The single bacterial time lapse images thus obtained are processed to determine the growth of bacteria in the presence of different concentrations of antibiotics. This method was used to determine the MIC values of 3 standard CLSI strains within 3-4 hours [26]. The results also exhibited good correlation between the MIC values obtained using the MAC system to that of the CLSI standards.
Although, this method was rapid for the strains tested in the article, the method is limited by the doubling time of the bacteria. For example, norflaxacin against S. aureus took 7 hours, as S. aureus tends to grow slower in its presence above MIC values. Thus, MIC values for slower growing organisms may potentially take much longer than 4 hours. It is also unclear if this method can be used directly from patient samples or needs pre-processing to isolate pure microbial cultures.
Surface enhanced raman spectroscopy-AST: This method uses a Surface Enhanced Raman Spectra (SERS) substrate based on two-dimensional hexagonally packed silver nanoparticles embedded in nano-channels of anodic aluminum oxide. This method is able to identify SERS spectroscopic biomarkers for determination of bacterial MIC against antibiotics. When bacteria in the sample is susceptible to the presence of an antibiotic at a given concentration, the SERS pattern decreases in amplitude over time, while the resistant strains do not show any significant change in their spectral pattern. The authors [27] were able to demonstrate the change in spectral pattern of S. aureus-oxacillin and E. coli-imipenem within 2 hours. Additionally, they determined the breakpoint value of spectral signal ratios, above which the antibiotic would be considered ineffective and below which they are considered susceptible. This breakpoint value was also used to determine the MIC values and showed good correlation with standard broth dilution method with no major errors. By optimizing the antibiotic treatment protocols and methodology, they may be able to bring down the treatment time to within an hour.
However, the breakpoint value currently identified in the article is based only on selected bacteria-antibiotic combinations and further studies with clinically relevant bacteria are required to either determine a new breakpoint or establish that the current breakpoint values are valid for all cases. Unlike several methods mentioned previously, this method requires pure bacterial cultures for AST determination which delays the time taken to obtain the AST results from patient samples.
FAS test (Fast AST method): This process uses microfluidic channels to trap bacterial cells within the channels, load them with media and monitor the cell growth with microscopic imaging to determine the effect of antibiotic on individual cells. The growth rate calculations are done for each individual cell traps in reference rows (which do not contain any antibiotics) and treatment rows (containing antibiotics at different concentrations). By averaging and normalizing the growth rate against reference population, they can detect the response to antibiotic treatment populations [28]. This method was used to determine the AST for E coli with respect to nine different antibiotics commonly used for Urinary Tract Infections (UTIs). They were able to expand the method to clinical isolates with high specificity for detecting resistance within 30 minutes.
However, since they observe for growth patterns among bacteria, the time taken for AST is dependent on the doubling time of the bacteria and hence can be high for slow growing bacteria. The method can directly use urine samples from patients for AST but may need pure cultures for other body fluids.
Isothermal micro calorimetry (IMC): IMC measures the heat flowrate of a given bacterial sample in suspension to determine the bacterial growth or lack thereof. Since heat produced is proportional to the reaction rate in the suspension, the heat signatures obtained by the bacterial suspension in presence or absence of antibiotics can be used to determine the effect of antibiotic on the bacteria and in turn determine MIC [29].
Apart from determining MIC, this method can also be used to distinguish between bacteriostatic vs bactericidal effect of the antibiotic on the bacteria. If the heat curves obtained produced a delay in onset of the growth they are termed bacteriostatic in nature. A detectable heat flow indicates presence of sufficient number of bacteria to produce significant heat signal, which implies ineffectiveness of antibiotic at a given concentration of bacteria. Based on the combination of heat flow and aggregate heat curves of antibiotic on the bacteria this method is also able to group the antibiotic into different modes of action such as inhibitors of cell wall synthesis, DNA or protein synthesis. It may be potentially a useful tool in determining and classifying new antibiotics and in drug discovery.
However, the time taken to obtain the MIC results is ~ 24 hours and hence may not be advantageous if faster AST results are required. There may be delays in onset of heat production due to low initial bacterial counts in the sample, which may lead to misinterpretation of mode of action or MIC determination.
Microfluidic electrical impedance spectroscopy (m- EIS) system
This method uses impedance measurements over a wide frequency range to determine the change in the capacitance of the suspension containing bacteria and antibiotics. In the presence of an Alternating Current (AC) electric field, the bacteria present in their specially designed microfluidic channels can store charge across their membrane and hence act as capacitors. As the number of bacteria in the channel increase, the capacitance of the suspension is said to increase, while as the bacteria die (above MIC values of antibiotics), they lose their membrane potential, and as a result, the capacitance of the suspension containing bacteria-antibiotic will decrease. This can be seen as a decrease in their electrical signal. Thus, the concentration of the antibiotic at which no change in the signal is seen over time or when a decrease in the signal is observed, is considered their MIC. Since the system does not need to wait to see bacterial growth-which is limited by the doubling time of the bacteria, but detect bacterial death in the presence of antibiotics, it can lead to faster AST profiles (<4 hours) [30]. The method can also distinguish between bactericidal and bacteriostatic effect of the antibiotics on samples and can go directly from samples without need for pure isolates. However, this system is in its early stages, with limited quality control strains tested against the specific antibiotics. Further testing with clinical isolates and automation of the method is required to determine its time savings.
Conclusion
The current systems in the market are considered gold standard and are highly reliable systems being used for decades in the clinical microbiology labs. However, they mostly rely on detecting changes based on bacterial metabolism and require pure cultural isolates. Due to these reasons, they take a long time to obtain antibiotic susceptibility profiles for pathogens, which in turn causes delay in providing appropriate targeted treatment. Hence, research is now shifting towards developing rapid AST systems which are able to bypass the need for pure clinical isolates. These novel systems can either directly or through simple pre-processing of samples be used for direct AST determination from patient samples or positive culture samples. The systems employ novel approaches for AST determination including use of microscopy, DNA probes, micro-cantilevers to name a few. Such novel devices will in the near future be able to replace the current gold standards, be faster and equally reliable in obtaining results. The new emerging technologies are also taking a similar path and are aimed at reducing the time taken between acquiring patient samples and reporting the susceptibility profiles for targeted treatment of patients. These technologies use innovative approaches such as microcalorimetry, impedance, Raman spectroscopy etc. to achieve their goals. These trends will inherently improve the turnaround times for sample processing in the labs, reduce the burden on technicians, provide rapid reporting of AST, with the ultimate goal of faster treatment of patients, reduced load on use of broad-spectrum antibiotics and better clinical outcomes.
Acknowledgement
We would like to thank Harold Burgeson, Lab supervisor and the staff at the clinical microbiology lab, University of Missouri Hospital, for their inputs.
Conflict of Interest
The authors Dr. Puttaswamy and Dr. Sengupta are coinventors of the m-EIS technology described under the future AST technologies in the article.
22801
References
- Andersson DI, Hughes D (2010) Antibiotic resistance and its cost: is it possible to reverse resistance? Nature Reviews Microbiology 8: 260.
- Neu HC (1992) The crisis in antibiotic resistance. Science 257: 1064-1073.
- Martínez JL (2008) Antibiotics and antibiotic resistance genes in natural environments. Science 321: 365-367.
- Organization WH (2017) Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. Geneva: World Health Organization 12: 154.
- Control CfD Prevention (2013) Antibiotic resistance threats in the United States. Centres for Disease Control and Prevention, US Department of Health and Human Services 124.
- O’Neill J (2014) Antimicrobial resistance: tackling a crisis for the health and wealth of nations. The Review on Antimicrobial Resistance 20.
- Zadroga R, Williams DN, Gottschall R, Hanson K, Nordberg V, et al. (2013) Comparison of 2 blood culture media shows significant differences in bacterial recovery for patients on antimicrobial therapy. Clinical Infectious Diseases 56: 790-797.
- Puskarich MA, Trzeciak S, Shapiro NI, Arnold RC, Horton JM, et al. (2011) Association between timing of antibiotic administration and mortality from septic shock in patients treated with a quantitative resuscitation protocol. Critical care medicine 39: 2066.
- Andrews JM (2001) Determination of minimum inhibitory concentrations. Journal of antimicrobial Chemotherapy 48: 5-16.
- MacGowan AP, Wise R (2001) Establishing MIC breakpoints and the interpretation of in vitro susceptibility tests. Journal of Antimicrobial Chemotherapy 48: 17-28.
- Marschal M, Bachmaier J, Autenrieth I, Oberhettinger P, Willmann M, et al. (2017) Evaluation of the Accelerate Pheno™ system for fast identification and antimicrobial susceptibility testing from positive blood culture in Gram-negative bloodstream infection. Journal of clinical microbiology: JCM 22: 27.
- Ferraro MJ (2000) Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. NCCLS 14: 13-19.
- Wellinghausen N, Pietzcker T, Poppert S, Belak S, Fieser N, et al. (2007) Evaluation of the Merlin MICRONAUT system for rapid direct susceptibility testing of gram-positive cocci and gram-negative bacilli from positive blood cultures. Journal of clinical microbiology 45:789-795.
- Reller LB, Weinstein M, Jorgensen JH, Ferraro MJ (2009) Antimicrobial susceptibility testing: a review of general principles and contemporary practices. Clinical infectious diseases 49: 1749-1755.
- White RL, Burgess DS, Manduru M, Bosso JA (1996) Comparison of three different in vitro methods of detecting synergy: time-kill, checkerboard, and E test. Antimicrobial agents and chemotherapy 40: 1914-1918.
- Ligozzi M, Bernini C, Bonora MG, de Fatima M, Zuliani J, et al. (2002) Evaluation of the VITEK 2 system for identification and antimicrobial susceptibility testing of medically relevant gram-positive cocci. Journal of clinical microbiology 40: 1681-1686.
- Bachmaier J, Autenrieth I (1998) Substantial equivalence determination decision summary; assay only template. Clinical Infectious Diseases 16: 79-87.
- Carroll KC, Glanz BD, Borek AP, Burger C, Bhally HS, et al. (2006) Evaluation of the BD Phoenix automated microbiology system for identification and antimicrobial susceptibility testing of Enterobacteriaceae. Journal of clinical microbiology 44: 3506-3509.
- Belak JM (2001) Determination of minimum inhibitory concentrations. Journal of antimicrobial Chemotherapy 48: 5-16.
- MicroScan WalkAway (1996) Substantial equivalence determination decision summary; assay only template. K041150.
- Hayden RT, Clinton LK, Hewitt C, Koyamatsu T, Sun Y, et al. (2016) Rapid antimicrobial susceptibility testing using forward laser light scatter technology. Journal of clinical microbiology 54: 2701-2706.
- Chantell C (2015) Multiplexed automated digital microscopy for rapid identification and antimicrobial susceptibility testing of bacteria and yeast directly from clinical samples. Clinical Microbiology Newsletter 37:161-167.
- Schneider CB, Ken Harris, Peter Khera, Kaveri Strenn (2001) Steve Rapid Antimicrobial Susceptibility Tests by Mass Measurement on a 96-Well Plate. Nature nanotechnology 18: 222.
- Longo G, Alonso-Sarduy L, Rio LM, Bizzini A, Trampuz A, et al. 2013. Rapid detection of bacterial resistance to antibiotics using AFM cantilevers as nanomechanical sensors. Nature nanotechnology 8: 522.
- Choi J, Jung Y-G, Kim J, Kim S, Jung Y, et al. (2013) Rapid antibiotic susceptibility testing by tracking single cell growth in a microfluidic agarose channel system. Lab on a Chip 13: 280-287.
- Liu CY, Han YY, Shih PH, Lian WN, Wang HH, et al. (2016) Rapid bacterial antibiotic susceptibility test based on simple surface-enhanced Raman spectroscopic biomarkers. Scientific reports 6: 23375.
- Baltekin O, Boucharin A, Tano E, Andersson DI, Elf J (2017) Antibiotic susceptibility testing in less than 30 min using direct single-cell imaging. Proceedings of the National Academy of Sciences 20: 558.
- Von Ah U, Wirz D, Daniels A (2009) Isothermal micro calorimetry–a new method for MIC determinations: results for 12 antibiotics and reference strains of E. coli and S. aureus. BMC microbiology 9: 106.
- Puttaswamy S, Lee B, Amighi B, Chakraborty S, Sengupta S (2012) Novel electrical method for the rapid determination of minimum inhibitory concentration (MIC) and assay of bactericidal/bacteriostatic activity. J Biosens Bioelectron S 2: 003.






