Keywords
Tigecycline; Susceptibility breakpoints; Antimicrobial resistance
Introduction
Tigecycline is the first commercially available member of the glycylcycline class of antimicrobials, and is approved in the USA for the treatment of complicated Skin and Skin Structure Infections (cSSSI), complicated Intra-abdominal Infections (cIAI) and Community-Acquired Bacterial Pneumonia (CAP) and for cSSSI and cIAIs in Europe [1,2]. Several phase III, and phase IIIb/IV, clinical trials have been conducted to establish the clinical efficacy of tigecycline in the treatment of cIAIs, cSSSIs and CAP and their data have been published previously [3–9]. Clinical trials have also been carried out for tigecycline in diabetic foot infection and nosocomial pneumonia; however, efficacy was not established in these trials [10-12]. The tigecycline susceptibility breakpoints as presented in the tigecycline package insert have been designated by the USA Food and Drug Administration (FDA) and are derived from clinical trial data of outcome in mono- and polymicrobial infection. The Clinical and Laboratory Standards Institute (CLSI) has not published susceptibility breakpoints for tigecycline as the data have not been made available by the sponsor. Susceptibility breakpoints for other antimicrobials published by CLSI are based on PharmacoKinetic/PharmacoDynamic (PK/PD) relationships and Minimum Inhibitory Concentration (MIC)/disk diffusion zone diameter distributions plus clinical outcome data [13]. European breakpoints for tigecycline have been made available by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) [14]. FDA and EUCAST breakpoints for tigecycline are shown in Table 1.
| Pathogen |
FDA |
EUCAST |
| |
S |
I |
R |
S |
R |
| Gram-negative |
|
|
|
|
|
| Enterobacteriaceae |
≤2 |
4 |
≥8 |
≤1 |
>2 |
| Acinetobacterbaumanniib |
- |
- |
- |
“IE” |
“IE” |
| Gram-positive |
|
|
|
|
|
| S. aureus |
≤0.5 |
- |
- |
≤0.5 |
>0.5 |
| S. pneumoniae |
≤0.06 |
- |
- |
“IE” |
“IE” |
| Streptococcus spp. (non-pneumoniae) |
≤0.06 |
- |
- |
≤0.25 |
>0.5 |
| E. faecalis |
≤0.25 |
- |
- |
≤0.25 |
>0.5 |
| E. faecium |
≤0.25 |
- |
- |
≤0.25 |
>0.5 |
a No CLSI breakpoints are available for tigecycline.
b No tigecycline breakpoints are provided by the FDA for A. baumannii; in this study, we use a susceptibility breakpoint of ≤2 mg/L as recommended by Jones et al. (25).
S = susceptible; I = intermediate; R = resistant; “IE” = “there is insufficient evidence that the species in question is a good target for therapy with the drug” (14).
Table 1: Tigecyclinea breakpoints (mg/L) for important Gram-negative and Gram-positive pathogens according to Food and Drug Administration (FDA) and European Committee on Antimicrobial Susceptibility Testing (EUCAST).
The PharmacoKinetics (PK) of tigecycline, after short IV infusions, are best described by a two-compartment model with zero-order input and linear distribution and elimination [15,16]. Maximum concentration and Area Under the Curve (AUC) are dose proportional after single doses of 12.5 to 300 mg and multiple doses of 25 to 100 mg q12h. Tigecycline has a large volume of distribution, ranging from 2.4 to 7 L/kg in healthy subjects [17], and has been shown to be extensively distributed in tissues. Interestingly, it exhibits non-linear plasma protein binding over therapeutic concentrations (the unbound fraction decreases as concentration increases) [18,19]. The rapid tissue penetration of tigecycline results in low serum concentrations, which may underestimate apparent drug concentrations in various tissues and may obscure the relationship between serum concentration and clinical outcome [17]. Tigecycline activity is time-dependent with an extended post-antibiotic effect, and is best predicted by the 24 h AUC:MIC ratio [20,21]. A long half-life (37–64 h) has been reported for tigecycline due to its slow release from tissues [16], although its accumulation is consistent with a shorter halflife [17]. Tigecycline demonstrates a mean clearance of 0.19 to 0.34 L/h/kg, with biliary elimination predominant [18].
The goal of this investigation is to evaluate the FDA approved breakpoints for tigecycline using microbiological and clinical data derived from cSSSI, cIAI and CAP clinical trials carried out both before and since the FDA breakpoints for tigecycline were established, as well as using microbiology data from the Tigecycline Evaluation and Susceptibility Trial (T.E.S.T.).
Materials and Methods
Microbiological eradication and clinical cure data for patients from the tigecycline phase III and IIIb/IV clinical trials for cSSSI, cIAI and CAP (the three indications for which tigecycline haS FDA approval) were obtained. Data from the clinical trials for tigecycline in diabetic foot infection and nosocomial pneumonia were not included. Populations used in this analysis include the Microbiological Modified Intent to Treat Population (M-MITT) and the Microbiologically Evaluable (ME) population. The M-MITT population was made up of patients who received at least one dose of study drug and had a baseline organism isolated. Details on the inclusion and exclusion criteria as well as other criteria used in the trials, along with population definitions, have been presented previously [3–9]. The primary analysis was carried out on the ME population and results are presented for this population.
In vitro activity data for tigecycline against organisms collected in the USA between 2004 and 2012 were collected from the T.E.S.T. database. In vitro testing methodologies, including determination of Minimum Inhibitory Concentrations (MICs), used in T.E.S.T. have been described previously [22,23]. The FDA-approved tigecycline breakpoints used in this study are shown in Table 1. Provisional tigecycline MIC breakpoints have been designated by Jones et al. [24] for Acinetobacter as susceptible, ≤2 mg/L; intermediate, 4 mg/L; and resistant, ≥8 mg/L (Table 1).
A Fisher’s exact test was used to compare microbiological eradication rates by MIC. The Cochran Armitage trend test was used to identify statistically significant changes in susceptibility between 2004 and 2012 among the T.E.S.T. data. A positive result indicated a statistically significant decrease in antimicrobial susceptibility. A minimum value of p<0.01 was used for statistical testing. p<0.05 was not used as computation of a high number of statistical tests can lead to significant results purely by chance, and use of a lower significance value reduces the possibility of this occurring.
Results
Microbiological and clinical success rates by indication, by key pathogen, and by MIC, for the ME population are presented in Table 2 and pooled data for Gram-negative and Gram-positive pathogens are presented in Table 3. The distribution of key pathogens by indication is presented in Figure 1. Microbiological eradication of Enterobacteriaceae was observed among 82.7%, 78.9% and 100% of microbiologically evaluable cIAI, cSSSI and CAP patients treated with tigecycline, respectively. The corresponding clinical cure rates among patients infected with Enterobacteriaceae in the cIAI, cSSSI and CAP treatment groups were 81.2%, 75.8% and 100%, respectively (Table 2).
| Pathogen |
MIC |
Microbiological eradication, n/N (%) |
Clinical Cure, n/N (%) |
| cIAI |
|
|
|
| Gram-negative |
|
|
|
| Enterobacteriaceaea |
≤0.06 |
28/35 (80.0) |
27/35 (77.1) |
| |
0.12 |
125/155 (80.6) |
122/155 (78.7) |
| |
0.25 |
278/329 (84.5) |
273/329 (83.0) |
| |
0.5 |
149/175 (85.1) |
147/175 (84.0) |
| |
1 |
50/62 (80.6) |
50/62 (80.6) |
| |
2 |
23/29 (79.3) |
22/29 (75.9) |
| |
4 |
6/11 (54.5) |
6/11 (54.5) |
| |
8 |
1/1 |
1/1 |
| |
16 |
0/1 |
0/1 |
| |
Total |
660/798 (82.7) |
648/798 (81.2) |
| Enterobacter spp. |
0.25 |
5/6 |
5/6 |
| |
0.5 |
16/18 (88.9) |
16/18 (88.9) |
| |
1 |
7/10 (70.0) |
7/10 (70.0) |
| |
Total |
28/34 (82.4) |
28/34 (82.4) |
| E. coli |
≤0.06 |
28/35 (80.0) |
27/35 (77.1) |
| |
0.12 |
121/147 (82.3) |
118/147 (80.3) |
| |
0.25 |
235/276 (85.1) |
234/276 (84.8) |
| |
0.5 |
64/77 (83.1) |
63/77 (81.8) |
| |
1 |
6/6 |
6/6 |
| |
Total |
454/541 (83.9) |
448/541 (82.8) |
| E. coli (ESBL-pos.) |
0.25 |
2/3 |
2/3 |
| |
0.5 |
3/5 |
3/5 |
| |
1 |
1/1 |
1/1 |
| |
Total |
6/9 |
6/9 |
| K. oxytoca |
0.25 |
9/10 (90.0) |
8/10 (80.0) |
| |
0.5 |
13/14 (92.9) |
13/14 (92.9) |
| |
Total |
22/24 (91.7) |
21/24 (87.5) |
| K. pneumoniae |
0.12 |
1/1 |
1/1 |
| |
0.25 |
11/18 (61.1) |
10/18 (55.6) |
| |
0.5 |
35/42 (83.3) |
34/42 (81.0) |
| |
1 |
22/26 (84.6) |
22/26 (84.6) |
| |
2 |
5/6 |
5/6 |
| |
4 |
1/1 |
1/1 |
| |
Total |
75/94 (79.8) |
73/94 (77.7) |
| K. pneumoniae (ESBL-pos.) |
0.25 |
2/2 |
2/2 |
| |
0.5 |
2/2 |
2/2 |
| |
1 |
2/3 |
2/3 |
| |
2 |
1/1 |
1/1 |
| |
Total |
7/8 |
7/8 |
| Serratia spp. |
0.25 |
1/1 |
1/1 |
| |
1 |
1/2 |
1/2 |
| |
2 |
3/3 |
3/3 |
| |
Total |
5/6 |
5/6 |
| Acinetobacter spp. |
≤0.06 |
1/1 |
1/1 |
| |
0.12 |
4/5 |
4/5 |
| |
0.25 |
3/4 |
3/4 |
| |
0.5 |
1/1 |
1/1 |
| |
1 |
4/4 |
4/4 |
| |
2 |
2/2 |
2/2 |
| |
Total |
15/17 (88.2) |
15/17 (88.2) |
| Gram-positive |
|
|
|
| E. faecalis |
0.03 |
2/2 |
2/2 |
| |
0.06 |
7/9 |
7/9 |
| |
0.12 |
16/22 (72.7) |
16/22 (72.7) |
| |
0.25 |
12/17 (70.6) |
10/17 (58.8) |
| |
Total |
37/50 (74.0) |
35/50 (70.0) |
| E. faecium |
0.015 |
0/1 |
0/1 |
| |
0.03 |
3/3 |
3/3 |
| |
0.06 |
6/8 |
6/8 |
| |
0.12 |
6/8 |
6/8 |
| |
0.25 |
2/2 |
2/2 |
| |
Total |
17/22 (77.3) |
17/22 (77.3) |
| S. aureus |
≤0.06 |
1/1 |
0/1 |
| |
0.12 |
20/21 (95.2) |
20/21 (95.2) |
| |
0.25 |
13/16 (81.3) |
13/16 (81.3) |
| |
0.5 |
1/1 |
1/1 |
| |
Total |
35/39 (89.7) |
34/39 (87.2) |
| MRSA |
0.12 |
1/1 |
1/1 |
| |
0.25 |
2/3 |
2/3 |
| |
Total |
3/4 |
3/4 |
| S. pneumoniae |
0.03 |
2/2 |
2/2 |
| |
0.06 |
1/1 |
1/1 |
| |
0.12 |
1/2 |
1/2 |
| |
Total |
4/5 |
4/5 |
| |
|
|
|
| cSSSI |
|
|
|
| Gram-negative |
|
|
|
| Enterobacteriaceaea |
0.12 |
7/9 |
7/9 |
| |
0.25 |
34/40 (85.0) |
34/40 (85.0) |
| |
0.5 |
30/39 (76.9) |
28/39 (71.8) |
| |
1 |
9/14 (64.3) |
8/14 (57.1) |
| |
2 |
18/23 (78.3) |
17/23 (73.9) |
| |
4 |
3/3 |
3/3 |
| |
Total |
101/128 (78.9) |
97/128 (75.8) |
| Enterobacter spp. |
0.25 |
5/6 |
5/6 |
| |
0.5 |
8/9 |
6/9 |
| |
1 |
4/6 |
3/6 |
| |
2 |
1/1 |
1/1 |
| |
Total |
18/22 (81.8) |
15/22 (68.2) |
| E. coli |
0.12 |
7/9 |
7/9 |
| |
0.25 |
22/25 (88.0) |
22/25 (88.0) |
| |
0.5 |
5/7 |
5/7 |
| |
1 |
1/1 |
1/1 |
| |
Total |
35/42 (83.3) |
35/42 (83.3) |
| E. coli (ESBL-pos.) |
0.25 |
1/1 |
0/1 |
| |
Total |
1/1 |
0/1 |
| K. oxytoca |
0.25 |
2/4 |
2/4 |
| |
0.5 |
3/5 |
2/5 |
| |
Total |
5/9 |
4/9 |
| K. pneumoniae |
0.25 |
1/1 |
1/1 |
| |
0.5 |
8/11 (72.7) |
10/11 (90.9) |
| |
2 |
2/2 |
2/2 |
| |
Total |
11/14 (78.6) |
13/14 (92.9) |
| K. pneumoniae (ESBL-pos.) |
- |
NA |
NA |
| Serratia spp. |
0.5 |
1/1 |
0/1 |
| |
1 |
2/3 |
2/3 |
| |
2 |
3/4 |
2/4 |
| |
Total |
6/8 |
4/8 |
| Acinetobacterspp. |
0.12 |
2/3 |
2/3 |
| |
0.25 |
2/4 |
2/4 |
| |
0.5 |
2/2 |
2/2 |
| |
1 |
1/2 |
1/2 |
| |
Total |
7/11 (63.6) |
7/11 (63.6) |
| Gram-positive |
|
|
|
| E. faecalis |
0.12 |
14/17 (82.4) |
13/17 (76.5) |
| |
0.25 |
5/5 |
3/5 |
| |
Total |
19/22 (86.4) |
16/22 (72.7) |
| E. faecium |
0.03 |
1/1 |
1/1 |
| |
0.06 |
5/5 |
5/5 |
| |
0.12 |
1/1 |
1/1 |
| |
0.5 |
0/1 |
0/1 |
| |
Total |
7/8 |
7/8 |
| S. aureus |
0.03 |
1/1 |
1/1 |
| |
0.06 |
7/9 |
7/9 |
| |
0.12 |
147/179 (82.1) |
151/179 (84.4) |
| |
0.25 |
40/46 (87.0) |
40/46 (87.0) |
| |
0.5 |
1/1 |
1/1 |
| |
1 |
1/1 |
1/1 |
| |
Total |
197/237 (83.1) |
201/237 (84.8) |
| MRSA |
0.06 |
4/6 |
4/6 |
| |
0.12 |
35/47 (74.5) |
35/47 (74.5) |
| |
0.25 |
10/14 (71.4) |
10/14 (71.4) |
| |
Total |
49/67 (73.1) |
49/67 (73.1) |
| S. pneumoniae |
0.03 |
1/1 |
1/1 |
| |
Total |
1/1 |
1/1 |
| |
|
|
|
| CAP |
|
|
|
| Gram-negative |
|
|
|
| Enterobacteriaceaea |
≤0.06 |
1/1 |
1/1 |
| |
0.12 |
2/2 |
2/2 |
| |
0.25 |
3/3 |
3/3 |
| |
0.5 |
3/3 |
3/3 |
| |
1 |
2/2 |
2/2 |
| |
2 |
1/1 |
1/1 |
| |
Total |
12/12 (100) |
12/12 (100) |
| Enterobacter spp. |
- |
NA |
NA |
| E. coli |
≤0.06 |
1/1 |
1/1 |
| |
0.12 |
2/2 |
2/2 |
| |
0.25 |
2/2 |
2/2 |
| |
Total |
5/5 |
5/5 |
| E. coli (ESBL-pos.) |
- |
NA |
NA |
| K. oxytoca |
0.5 |
1/1 |
1/1 |
| |
Total |
1/1 |
1/1 |
| K. pneumoniae |
0.5 |
2/2 |
2/2 |
| |
1 |
2/2 |
2/2 |
| |
Total |
4/4 |
4/4 |
| K. pneumoniae (ESBL-pos.) |
- |
NA |
NA |
| Serratia spp. |
2 |
1/1 |
1/1 |
| |
Total |
1/1 |
1/1 |
| Acinetobacter spp. |
0.12 |
1/2 |
1/2 |
| |
Total |
1/2 |
1/2 |
| Gram-positive |
|
|
|
| E. faecalis |
- |
NA |
NA |
| E. faecium |
- |
NA |
NA |
| S. aureus |
0.12 |
7/9 |
7/9 |
| |
0.25 |
2/3 |
2/3 |
| |
Total |
9/12 (75.0) |
9/12 (75.0) |
| MRSA |
- |
NA |
NA |
| S. pneumoniae |
0.03 |
4/4 |
4/4 |
| |
0.06 |
46/47 (97.9) |
46/47 (97.9) |
| |
0.12 |
1/2 |
0/2 |
| |
Total |
51/53 (96.2) |
50/53 (94.3) |
| |
|
|
|
| Combined |
|
|
|
| Gram-negative |
|
|
|
| Enterobacteriaceaea |
≤0.06 |
29/36 (80.6) |
28/36 (77.8) |
| |
0.12 |
134/166 (80.7) |
131/166 (78.9) |
| |
0.25 |
315/372 (84.7) |
310/372 (83.3) |
| |
0.5 |
182/217 (83.9) |
178/217 (82.0) |
| |
1 |
61/78 (78.2) |
60/78 (76.9) |
| |
2 |
42/53 (79.2) |
40/53 (75.5) |
| |
4 |
9/14 (64.3) |
9/14 (64.3) |
| |
8 |
1/1 |
1/1 |
| |
16 |
0/1 |
0/1 |
| |
Total |
773/938 (82.4) |
757/938 (80.7) |
| Enterobacter spp. |
0.25 |
10/12 (83.3) |
10/12 (83.3) |
| |
0.5 |
24/27 (88.9) |
22/27 (81.5) |
| |
1 |
11/16 (68.8) |
10/16 (62.5) |
| |
2 |
1/1 |
1/1 |
| |
Total |
46/56 (82.1) |
43/56 (76.8) |
| E. coli |
≤0.06 |
29/36 (80.6) |
28/36 (77.8) |
| |
0.12 |
130/158 (82.3) |
127/158 (80.4) |
| |
0.25 |
259/303 (85.5) |
258/303 (85.1) |
| |
0.5 |
69/84 (82.1) |
68/84 (81.0) |
| |
1 |
7/7 |
7/7 |
| |
Total |
494/588 (84.0) |
488/588 (83.0) |
| E. coli (ESBL-pos.) |
0.25 |
3/4 |
2/4 |
| |
0.5 |
3/5 |
3/5 |
| |
1 |
1/1 |
1/1 |
| |
Total |
7/10 (70.0) |
6/10 (60.0) |
| K. oxytoca |
0.25 |
11/14 (78.6) |
10/14 (71.4) |
| |
0.5 |
17/20 (85.0) |
16/20 (80.0) |
| |
Total |
28/34 (82.4) |
26/34 (76.5) |
| K. pneumoniae |
0.12 |
1/1 |
1/1 |
| |
0.25 |
12/19 (63.2) |
11/19 (57.9) |
| |
0.5 |
45/55 (81.8) |
46/55 (83.6) |
| |
1 |
24/28 (85.7) |
24/28 (85.7) |
| |
2 |
7/8 |
7/8 |
| |
4 |
1/1 |
1/1 |
| |
Total |
90/112 (80.4) |
90/112 (80.4) |
| K. pneumoniae (ESBL-pos.) |
0.25 |
2/2 |
2/2 |
| |
0.5 |
2/2 |
2/2 |
| |
1 |
2/3 |
2/3 |
| |
2 |
1/1 |
1/1 |
| |
Total |
7/8 |
7/8 |
| Serratia spp. |
0.25 |
1/1 |
1/1 |
| |
0.5 |
1/1 |
0/1 |
| |
1 |
3/5 |
3/5 |
| |
2 |
7/8 |
6/8 |
| |
Total |
12/15 (80.0) |
10/15 (66.7) |
| Acinetobacter spp. |
≤0.06 |
1/1 |
1/1 |
| |
0.12 |
7/10 (70.0) |
7/10 (70.0) |
| |
0.25 |
5/8 |
5/8 |
| |
0.5 |
3/3 |
3/3 |
| |
1 |
5/6 |
5/6 |
| |
2 |
2/2 |
2/2 |
| |
Total |
23/30 (76.7) |
23/30 (76.7) |
| Gram-positive |
|
|
|
| E. faecalis |
0.03 |
2/2 |
2/2 |
| |
0.06 |
7/9 |
7/9 |
| |
0.12 |
30/39 (76.9) |
29/39 (74.4) |
| |
0.25 |
17/22 (77.3) |
13/22 (59.1) |
| |
Total |
56/72 (77.8) |
51/72 (70.8) |
| E. faecium |
0.015 |
0/1 |
0/1 |
| |
0.03 |
4/4 |
4/4 |
| |
0.06 |
11/13 (84.6) |
11/13 (84.6) |
| |
0.12 |
7/9 |
7/9 |
| |
0.25 |
2/2 |
2/2 |
| |
0.5 |
0/1 |
0/1 |
| |
Total |
24/30 (80.0) |
24/30 (80.0) |
| S. aureus |
0.03 |
1/1 |
1/1 |
| |
0.06 |
8/10 (80.0) |
7/10 (70.0) |
| |
0.12 |
174/209 (83.3) |
178/209 (85.2) |
| |
0.25 |
55/65 (84.6) |
55/65 (84.6) |
| |
0.5 |
2/2 |
2/2 |
| |
1 |
1/1 |
1/1 |
| |
Total |
241/288 (83.7) |
244/288 (84.7) |
| MRSA |
0.06 |
4/6 |
4/6 |
| |
0.12 |
36/48 (75.0) |
36/48 ()75.0 |
| |
0.25 |
12/17 (70.6) |
12/17 (70.6) |
| |
Total |
52/71 (73.2) |
52/71 (73.2) |
| S. pneumoniae |
0.03 |
7/7 |
7/7 |
| |
0.06 |
47/48 (97.9) |
47/48 (97.9) |
| |
0.12 |
2/4 |
1/4 |
| |
Total |
56/59 (94.9) |
55/59 (93.2) |
| |
NA, not available
Percentages only calculated when n≥10
a: Enterobacteriaceae Group includes the following pathogens (Note: not all were present in this database): Alishewanella, Alterococcus, Aquamonas, Aranicola, Arsenophonus, Azotivirga, Blochmannia, Brenneria, Buchnera, Budvicia, Buttiauxella, Cedecea, Citrobacter, Cronobacter, Dickeya, Edwardsiella, Enterobacter, Erwinia, Escherichia, Ewingella, Grimontella, Hafnia, Klebsiella, Kluyvera, Leclercia, Leminorella, Moellerella, Morganella, Obesumbacterium, Pantoea, Pectobacterium, Candidatus Phlomobacter, Photorhabdus, Plesiomonas, Pragia, Proteus, Providencia, Rahnella, Raoultella, Salmonella, Samsonia, Serratia, Shigella, Sodalis, Tatumella, Trabulsiella, Wigglesworthia, Xenorhabdus, Yersinia, Yokenella
Table 2: Microbiological and clinical success rates for tigecycline by infection type, pathogen, and MIC (mg/L) among microbiologically evaluable, tigecycline-treated patients from tigecycline phase and IIIb/IV with complicated Intra-Abdominal Infection (cIAI), complicated Skin And Skin Structure Infection (cSSSI) or Community-Acquired Pneumonia (CAP) clinical trials.
| MIC |
Microbiological eradication, n/N (%) |
Clinical Cure, n/N (%) |
| Gram-negativesa |
|
|
| ≤0.06 |
30/37 (81.1%) |
29/37 (78.4%) |
| 0.12 |
141/176 (80.1%) |
138/176 (78.4%) |
| 0.25 |
320/380 (84.2%) |
315/380 (82.9%) |
| 0.5 |
185/220 (84.1%) |
181/220 (82.3%) |
| 1 |
66/84 (78.6%) |
65/84 (77.4%) |
| 2 |
44/55 (80.0%) |
42/55 (76.4%) |
| 4 |
9/14 (64.3%) |
9/14 (64.3%) |
| 8 |
1/1 (100%) |
1/1 (100%) |
| 16 |
0/1 (0%) |
0/1 (0%) |
| Total |
796/968 (82.2%) |
780/968 (80.6%) |
| Gram-positivesb |
|
|
| ≤0.06 |
87/95 (91.6%) |
86/95 (90.5%) |
| 0.12 |
213/261 (81.6%) |
215/261 (82.4%) |
| 0.25 |
74/89 (83.1%) |
70/89 (78.7%) |
| 0.5 |
2/3 (66.7%) |
2/3 (66.7%) |
| 1 |
1/1 (100%) |
1/1 (100%) |
| Total |
377/449 (84.0%) |
374/449 (83.3%) |
| Combined |
|
|
| ≤0.06 |
117/132 (88.6%) |
115/132 (87.1%) |
| 0.12 |
354/437 (81.0%) |
353/437 (80.8%) |
| 0.25 |
394/469 (84.0%) |
385/469 (82.1%) |
| 0.5 |
187/223 (83.9%) |
183/223 (82.1%) |
| 1 |
67/85 (78.8%) |
66/85 (77.6%) |
| 2 |
44/55 (80.0%) |
42/55 (76.4%) |
| 4 |
9/14 (64.3%) |
9/14 (64.3%) |
| 8 |
1/1 (100%) |
1/1 (100%) |
| 16 |
0/1 (0%) |
0/1 (0%) |
| Total |
1173/1417 (82.8%) |
1154/1417 (81.4%) |
a Gram-negative organisms include Enterobacteriaceae (see Table 2 Footnote) and Acinetobacter spp.
b Gram-positive organisms include E. faecalis, E. faecium, S. aureus, and S. pneumoniae
Table 3: Microbiological and clinical success rates for tigecycline against cumulative Gram-negative and Gram-positive pathogens by MIC (mg/L) among microbiologically evaluable tigecycline-treated patients from tigecycline phase and IIIb/IV complicated Intra-Abdominal Infection (cIAI), complicated Skin and Skin Structure Infection (cSSSI) and Community-Acquired Pneumonia (CAP) clinical trials.
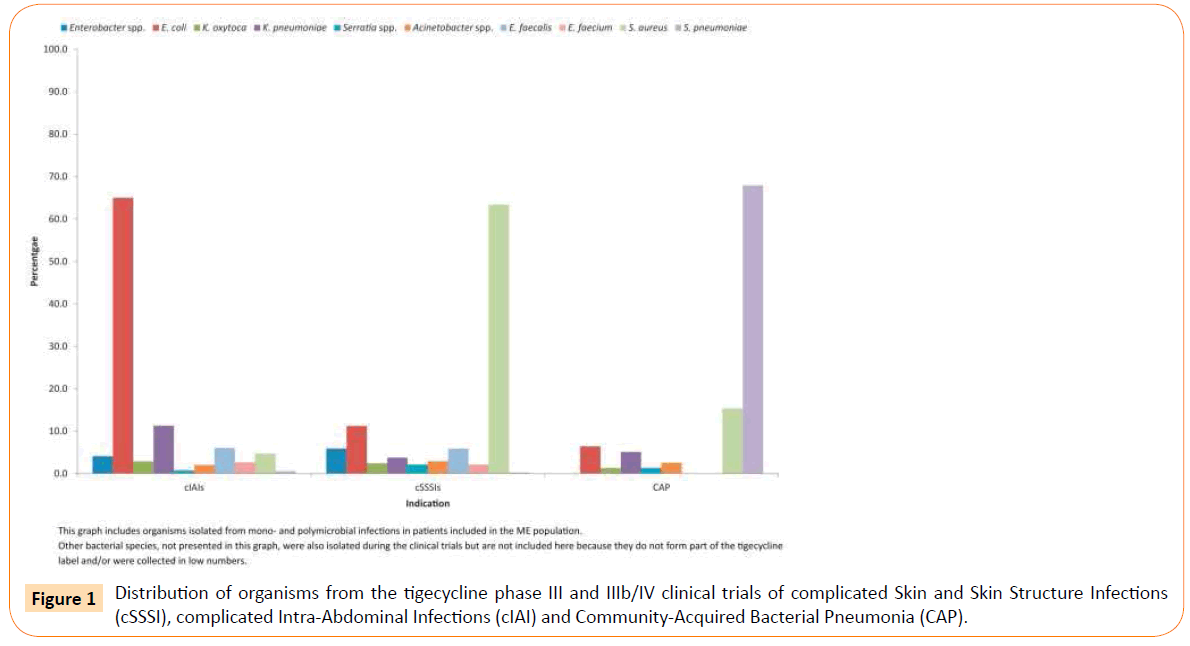
Figure 1: Distribution of organisms from the tigecycline phase III and IIIb/IV clinical trials of complicated Skin and Skin Structure Infections (cSSSI), complicated Intra-Abdominal Infections (cIAI) and Community-Acquired Bacterial Pneumonia (CAP).
Tigecycline non-susceptibility (MIC ≥4 mg/L) was observed among 16 Enterobacteriaceae isolates (1.7%), 13 of which were collected from patients with IAIs and three from patients with cSSSIs. These Enterobacteriaceae included a single isolate of K. pneumoniae as well as 12 Proteus spp. and 3 Morganella spp. Among Gram-positive pathogens, non-susceptibility was detected among a single isolate each of E. faecium (3.3%) and S. aureus (0.3%) as well as four S. pneumoniae isolates (6.8%). However, microbiological and clinical successes were reported for tigecycline against 13/22 (59.1%) and 12/22 (54.5%) of nonsusceptible pathogens, respectively. Of these 22 tigecycline nonsusceptible isolates, 16 were collected from cIAI patients.
Efficacy data for key pathogens in the tigecycline cIAI, cSSSI and CAP clinical trials have been summarized by MIC in Table 3. Microbiological eradication and clinical cure for tigecycline was observed among patients infected with pathogens with MICs up to 8 mg/L. Microbiological eradication was observed among 82.2% of patients infected with Gram-negative isolates and 84.0% infected with Gram-positive isolates; clinical cure was observed among 80.6% of patients infected with Gram-negative pathogens and among 83.3% infected with Gram-positive pathogens. The MIC of the majority of Enterobacteriaceae collected was ≤2 mg/L (the FDA susceptibility breakpoint for Enterobacteriaceae) and microbiological eradiation and clinical cure rates for patients from the three indications combined with infections caused by Enterobacteriaceae at an MIC of ≤2 mg/L were 82.8% (763/922) and 81.0% (747/922), respectively. No Gram-positive pathogens with MICs for tigecycline of greater than 1 mg/L were observed in the cIAI, cSSSI and CAP clinical trials.
When microbiological success rates for ME population patients with Enterobacteriaceae isolates with a MIC of ≤2 mg/L and ≥4 mg/L were compared (n=922 and n=16, respectively), the Fisher’s exact test produced a significant two-tailed P value of 0.046. With breakpoints of ≤4 and ≥8 mg/L, no significant difference was seen between microbiological success rates (P=0.321) although it should be noted that the n value in the ≥8 mg/L group was 2. There were 936 patients in the ≤4 mg/L group. For the M-MITT population, a non-significant two-tailed P value was obtained when comparing ≤2 mg/L and ≥4 mg/L isolates (P=0.431; n=1087 and n=20, respectively) and when comparing ≤4 and ≥8 mg/L (P=0.409). Again the n value in the ≥8 mg/L group was 2. There were 1105 patients in the ≤4 mg/L group.
For patients with S. aureus isolates comparisons of microbiological eradication rates by tigecycline MIC are difficult due to the narrow MIC range. Only one patient (who was in both the ME and M-MITT populations) was identified with an infection due to S. aureus with a tigecycline MIC of ≥1 mg/L. Comparing MICs of ≤0.5/≥1 mg/L (n=287 and n=1, respectively) in the ME population there were no statistically significant differences in microbiological success rates (P=1.000). The same result was gained for the M-MITT population (n=377 and n=1, respectively). No further comparisons between MIC groups could be made as there were no isolates with a tigecycline MIC of >1 mg/L.
Tigecycline in vitro potency data for key pathogens collected from patients in the USA between 2004 and 2012 through the T.E.S.T program are summarized and presented in Table 4. Tigecycline susceptibility did not change significantly during the study period for most pathogens (E. coli, K. oxytoca, K. pneumoniae, Serratia spp., S. aureus [including methicillin-resistant S. aureus (MRSA)], S. pneumoniae and E. faecalis). Statistically significant increases in tigecycline susceptibility were observed among Enterobacter spp. (p<0.01) and the Enterobacteriaceae (p<0.01). A statistically significant decrease in susceptibility was observed among E. faecium isolates (p<0.0001), representing a decrease from 100% in 2004 (n=243) to 96.9% in 2011 (n=97) before increasing again to 98.0% in 2012 (n=199). A small, statistically non-significant decrease in tigecycline susceptibility (1.9%) was also noted among Acinetobacter baumannii (using a tigecycline Enterobacteriaceae susceptibility breakpoint of ≤2 mg/L) (data not shown).
| Pathogen |
n |
MIC90 |
%S |
%S changea |
| Gram-negative |
|
|
|
|
| Enterobacteriaceae |
36 154 |
1 |
96.8 |
p<0.01 (+) |
| Enterobacter spp. |
9979 |
2 |
94.7 |
p<0.01 (+) |
| E. coli |
11 164 |
0.25 |
>99.9 |
NS |
| E. coli, ESBL-pos |
491 |
0.5 |
100 |
- |
| E. coli, ESBL-neg |
10 673 |
0.25 |
>99.9 |
NS |
| K. oxytoca |
1879 |
1 |
98.8 |
NS |
| K. oxytoca, ESBL-pos |
63 |
1 |
95.2 |
NS |
| K. oxytoca, ESBL-neg |
1816 |
1 |
98.9 |
NS |
| K. pneumoniae |
9004 |
2 |
95.2 |
NS |
| K. pneumoniae, ESBL-pos |
889 |
2 |
92.1 |
NS |
| K. pneumoniae, ESBL-neg |
8115 |
2 |
95.5 |
NS |
| Serratia spp. |
4128 |
2 |
96.3 |
NS |
| Acinetobacterbaumanniib |
4849 |
2 |
97.2 |
NS |
| Gram-positive |
|
|
|
|
| E. faecalis |
4658 |
0.25 |
99.5 |
NS |
| E. faecium |
1834 |
0.12 |
99.4 |
p<0.0001 (-) |
| S. aureus |
11 394 |
0.25 |
99.9 |
NS |
| S. aureus, MRSA |
6220 |
0.25 |
99.9 |
NS |
| S. aureus, MSSA |
5174 |
0.25 |
100 |
- |
| S. pneumoniae |
5358 |
0.06 |
99.8 |
NS |
a Cochrane Armitage trend test, comparing susceptible to non-susceptible (intermediate + resistant). p<0.01 was considered significant; a positive (+) trend test indicates significantly increased susceptibility whereas a negative (-) trend indicates significantly increased non-susceptibility
b Tigecycline is not indicated for the treatment of infections caused by A. baumannii. An Enterobacteriaceae susceptibility breakpoint of ≤2 mg/L was applied for A. baumannii as recommended by Jones et al. (25).
Table 4: Tigecycline minimum inhibitory concentration (MIC90 [mg/L]), antimicrobial susceptibility (%S), and statistically significant changes in susceptibilitya among pathogens collected from all patients in the USA during T.E.S.T 2004-2012.
Discussion
Our findings suggest that the susceptibility breakpoints assigned for tigecycline by the FDA are suitable for clinical use. In vitro data collected since the initial approval of tigecycline show that the phenotypic profile remains essentially unchanged. Breakpoints derived from clinical data, supported by in vitro data, may be more useful in predicting the effective treatment of infections than breakpoints derived from purely PK/PD considerations or epidemiological analysis, which may underestimate clinical efficacy.
The data presented in this report suggest a high level of agreement between microbiological eradication and clinical cure among patients treated with tigecycline in phase III and IIIb/IV trials for cSSSI, cIAI and CAP, although it should be noted that microbiological eradication is typically presumptive and not confirmed by retesting. Further, the data presented here indicate that there has been almost no decrease in tigecycline susceptibility in the USA between 2004 and 2012. Cochrane Armitage trend tests carried out on all Enterobacteriaceae (including Enterobacter spp., E. coli, Klebsiella spp., Serratia spp.), A. baumannii, Enterococcus spp., S. aureus and S. pneumoniae isolates collected during the study interval revealed that tigecycline susceptibility among the Enterobacteriaceae and Enterobacter spp. increased significantly (p<0.01) while nonsusceptibility increased significantly among E. faecium (p<0.0001) and no other significant changes in susceptibility were observed. The high correlation between microbiological and clinical success rates for tigecycline coupled with the prolonged stability of pathogen susceptibility to tigecycline suggests that the current tigecycline breakpoints remain appropriate for microbiological and clinical use.
Previous reports have shown that antimicrobial therapy can result in clinical success against infections caused by pathogens with MICs above CLSI or FDA approved breakpoints. In a study of moxifloxacin treatment of cIAIs caused by aerobic and anaerobic pathogens, Goldstein et al. found that clinical success was observed among pathogens with MICs higher than the susceptibility breakpoint of 2 mg/L [25]. Among infections caused by anaerobes susceptible to moxifloxacin at ≤2 mg/L, the clinical success rate was 83.1%; high clinical success rates were also reported at MICs of 4 mg/L (91.2%), 8 mg/L (82.4%) and 16 mg/L (83.3%). Bacteriological eradication at ≤2 mg/L was 84.5%, and remained high (91.2%) at 4 mg/L. In such cases it is also important to consider the occurrence of natural resolution and that clinical success can be observed above the resistance breakpoint because of this natural response; this may also be amplified in the treatment of conditions where surgical intervention plays a major role. In the current study few isolates of Enterobacteriaceae or S. aureus were collected with tigecycline MICs above the susceptibility breakpoints. However, the microbiological activity of tigecycline against the Enterobacteriaceae extended beyond the FDA-approved susceptibility breakpoints.
Ambrose et al. examined potential tigecycline breakpoints for Enterobacteriaceae using both calculation of clinical response expectation and the proportion of patients achieving a target PK/PD threshold (AUCss(0-24)/MIC ≥6.96) by means of Monte Carlo simulations [26]. These simulations provided similar results at MICs up to 0.12 mg/L, but the probability of PK/ PD Target Attainment (PTA) and clinical response expectation diverged at higher MIC values: the PK/PD target attainment model dramatically underestimated the clinical performance of tigecycline against Enterobacteriaceae at MICs ≥1 mg/L. This was due to the model assumption that only patients who attain the target PK/PD measure would be cured, without consideration of other potentially important factors such as surgical interventions or patient-specific co-variates such as APACHE II scores. This result is mirrored in the current study, in which successful clinical responses for infections caused by E. coli and S. aureus exceeded the PTA based on PK/PD derived breakpoints.
Tigecycline has previously demonstrated efficacy in both in vivo and in vitro studies where free serum concentrations remained below the MIC during the study interval. Crandon et al. showed Colony-Forming Unit (CFU) reductions among S. aureus isolates in a neutropenic murine thigh model study where tigecycline concentrations remained below the MIC during the complete study period, at times approaching maximum effect values for tigecycline versus S. aureus [27]. Similar results were reported by Scheetz et al. for tigecycline against carbapenem-intermediate and -resistant A. baumannii in vitro, with tigecycline concentrations of 0.7, 0.8, 0.9 and 1 mg/L producing similar activity to tigecycline at 2 mg/L (approximately 3-log10 decrease in 24 hours and 4-log10 decrease in 48 hours) [28]. These studies highlight the difficulties in describing the basis for clinical response in a drug such as tigecycline, where low serum concentrations and nonlinear protein binding can result in antimicrobial efficacy even at serum concentrations below MIC. In such cases, clinical results are more likely to be informative to clinicians than PK/PD results.
Antimicrobials are considered bactericidal if they achieve a 3-log10 kill over 24 hours, and so tigecycline is bacteriostatic in its activity (although bactericidal activity has been reported against Streptococcus pneumoniae and Legionella pneumophila) [1]. Tessier and Nicolau examined the activity of tigecycline against ESBL-producing E. coli and K. pneumoniae in a murine thigh infection model designed to mimic human dosing (100- mg loading dose followed by 50-mg doses q12h) [29]. In that study a 3-log10 kill was observed over 72 hours and the authors concluded that bactericidal activity was demonstrated in the setting of in vivo human experience.
Infections caused by carbapenemase producing Enterobacteriaceae, such as K. pneumoniae, are particularly challenging to treat. Tigecycline is considered a treatment option against these pathogens and one limitation of this study is the lack of data against such carbapenemase producing isolates.
Conclusion
The current FDA clinical breakpoints for tigecycline can be considered appropriate. Breakpoints derived from in vitro data supported by clinical trial results may be more meaningful in the treatment of patients with tigecycline-indicated infections than those breakpoints derived from pharmacokinetic/ pharmacodynamic data.
Acknowledgements
The authors thank all the clinical trial investigators and the many T.E.S.T. investigators and laboratories, as well as the staff at IHMA for their coordination of T.E.S.T. Thanks are also extended to the statistical analysis team at Pfizer Inc. Drs. Rod Taylor and Wendy Hartley (Micron Research Ltd, Ely, UK) provided editorial assistance, which was funded by Pfizer Inc. Micron Research Ltd also provided data management services which were funded by Pfizer Inc.
Funding
The clinical trials were funded by Wyeth Pharmaceuticals, now owned by Pfizer Inc. T.E.S.T. is funded by Pfizer Inc. As authors of the article include employees of Pfizer Inc, Pfizer Inc was involved in study design, analysis and interpretation of data; in writing the report; and in the decision to submit the article for publication.
Competing interests
David P. Nicolau has received research grants, participated in Speakers’ Bureau and acted as a consultant for Pfizer, Inc. Dr Nicolau received no funding for his involvement in the analysis, review and authoring of this manuscript. Michael J. Dowzicky, Joan Korth- Bradley, Alvaro Quintana and Michele Wible are employees of Pfizer Inc.
7474
References
- https://www.ema.europa.eu/docs/en_GB/document_library/EPAR_- Product_Information/human/000644/WC500044508.pdf
- Babinchak T, Ellis-Grosse E, Dartois N, Rose GM, Loh E, et al. (2005) The efficacy and safety of tigecycline for the treatment of complicated intra-abdominal infections: analysis of pooled clinical trial data. Clin Infect Dis 41: S354-S367.
- Ellis-Grosse EJ, Babinchak T, Dartois N, Rose G, Loh E, et al. (2005) The efficacy and safety of tigecycline in the treatment of skin and skin-structure infections: results of 2 double-blind phase 3 comparison studies with vancomycin-aztreonam. Clin Infect Dis 41: S341-S353.
- Tanaseanu C, Bergallo C, Teglia O, Jasovich A, Oliva ME, et al. (2008) Integrated results of 2 phase 3 studies comparing tigecycline and levofloxacin in community-acquired pneumonia. DiagnMicrobiol Infect Dis 61: 329-338.
- Chen Z, Wu J, Zhang Y, Wei J, Leng X, et al. (2010) Efficacy and safety of tigecyclinemonotherapy vs. imipenem/cilastatin in Chinese patients with complicated intra-abdominal infections: a randomized controlled trial. BMC Infect Dis 10: 217.
- Towfigh S, Pasternak J, Poirier A, Leister H, Babinchak T (2010) A multicentre, open-label, randomized comparative study of tigecycline versus ceftriaxone sodium plus metronidazole for the treatment of hospitalized subjects with complicated intra-abdominal infections. ClinMicrobiol Infect 16: 1274-1281.
- Qvist N, Warren B, Leister-Tebbe H, Zito ET, Pedersen R, et al. (2012) Efficacy of tigecycline versus ceftriaxone plus metronidazole for the treatment of complicated intra-abdominal infections: results from a randomized, controlled trial. Surg Infect 13: 102-109.
- Matthews P, Alpert M, Rahav G, Rill D, Zito E, et al. (2012) A randomized trial of tigecycline versus ampicillin-sulbactam or amoxicillin-clavulanate for the treatment of complicated skin and skin structure infections. BMC Infect Dis 12: 297.
- Freire AT, Melnyk V, Kim MJ, Datsenko O, Dzyublik O, et al. (2010) Comparison of tigecycline with imipenem/cilastatin for the treatment of hospital-acquired pneumonia. DiagnMicrobiol Infect Dis 68: 140-151.
- Lauf L, Ozsvár Z, Mitha I, Regöly-Mérei J, Embil JM, et al. (2014) Phase 3 study comparing tigecycline and ertapenem in patients with diabetic foot infections with and without osteomyelitis. DiagnMicrobiol Infect Dis 78: 469-480.
- Ramirez J, Dartois N, Gandjini H, Yan JL, Korth-Bradley J, et al. (2013) Randomized phase 2 trial to evaluate the clinical efficacy of two high-dosage tigecycline regimens versus imipenem-cilastatin for treatment of hospital-acquired pneumonia. Antimicrob Agents Chemother 57: 1756-1762.
- Jenkins SG, Jerris RC (2011) Critical assessment of issues applicable to development of antimicrobial susceptibility testing breakpoints. J ClinMicrobiol 49: S5-S10.
- www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_5.0_Breakpoint_Table_01.pdf
- Meagher A, Ambrose PG, Grasela TH, Ellis-Grosse EJ (2005) Thepharmacokinetic and pharmacodynamic profile of tigecycline. Clin Infect Dis 41: S333-S340.
- Van Wart SA, Owen JS, Ludwig EA, Meagher AK, Korth-Bradley JM, et al. (2006) Population pharmacokinetics of tigecycline in patients with complicated intra-abdominal or skin and skin structure infections. Antimicrob Agents Chemother 50:3701-3707.
- Muralidharan G, Micalizzi M, Speth J, Raible D, Troy S (2005)Pharmacokinetics of tigecycline after single and multiple doses in healthy subjects. Antimicrob Agents Chemother 49: 220-229.
- Estes KS, Derendorf H (2010) Comparison of the pharmacokinetic properties of vancomycin, linezolid, tigecycline, and daptomycin. Eur J Med Res 15: 533-543.
- Giamarellou H, Poulakou G (2011) Pharmacokinetic and pharmacodynamic evaluation of tigecycline. Expert Opin Drug MetabToxicol 7: 1459-1470.
- Agwuh KN, MacGowan A (2006) Pharmacokinetics and pharmacodynamics of the tetracyclines including glycylcyclines. J AntimicrobChemother 58: 256-265.
- Stein GE, Craig WA (2006) Tigecycline: a critical analysis. Clin Infect Dis 43: 518-424.
- Reinert RR, Low DE, Rossi F, Zhang X, Wattal C, et al. (2007) Antimicrobial susceptibility among organisms from the Asia/Pacific Rim, Europe and Latin and North America collected as part of TEST and the in vitro activity of tigecycline. J AntimicrobChemother 60: 1018-1029.
- Stefani S, Dowzicky MJ (2013) Longitudinal assessment of antimicrobial susceptibility among Gram-negative and Gram-positive organisms collected from Italy as part of the Tigecycline Evaluation and Surveillance Trial between 2004 and 2011. Pharmaceuticals Basel 6: 1381-1406.
- Jones RN, Ferraro MJ, Reller LB, Schreckenberger PC, Swenson JM, et al. (2007) Multicenter studies of tigecycline disk diffusion susceptibility results for Acinetobacter spp. J ClinMicrobiol 45: 2272-2230.
- Goldstein EJ, Solomkin JS, Citron DM, Alder JD (2011) Clinical efficacy and correlation of clinical outcomes with in vitro susceptibility for anaerobic bacteria in patients with complicated intra-abdominal infections treated with moxifloxacin. Clin Infect Dis 53: 1074-1080.
- Ambrose PG, Meagher AK, Passarell JA, Van Wart SA, Cirincione BB, et al. (2009) Use of a clinically derived exposure-response relationship to evaluate potential tigecycline-Enterobacteriaceae susceptibility breakpoints. DiagnMicrobiol Infect Dis 63: 38-42.
- Crandon JL, Banevicius MA, Nicolau DP (2009) Pharmacodynamics of tigecycline against phenotypically diverse Staphylococcus aureus isolates in a murine thigh model. Antimicrob Agents Chemother 53: 1165-1169.
- Scheetz MH, Qi C, Warren JR, Postelnick MJ, Zembower T, et al. (2007) In vitro activities of various antimicrobials alone and in combination with tigecycline against carbapenem-intermediate or -resistant Acinetobacterbaumannii. Antimicrob Agents Chemother 51: 1621-1626.
- Tessier PR, Nicolau DP (2013)Tigecycline displays in vivo bactericidal activity against extended-spectrum-ß-lactamase-producing Enterobacteriaceae after 72-hour exposure period. Antimicrob Agents Chemother 57: 640-642.






