Keywords
Leptospirosis; Jaundice-hemorrhage; Pulmonary hemorrhage; Leptospira interrogans; Serovar copenhageni
Abbreviations
ABG: Arterial Blood Gas; ICU: Intensive Care Unit; CT: Computed Tomography; MAT: Microscopic Agglutination Test; ELISA: Enzyme-Linked Immunosorbent Assay; PCR: Polymerase Chain Reaction.
Introduction
Leptospirosis is a fatal zoonotic infection occurs worldwide, especially in tropic region [1]. The symptoms of leptospirosis range from the common nonspecific flu-like illness to severe jaundice, renal failure, pneumorrhagia or meningitis. Severe manifestations are characterized by jaundice, acute renal failure and hemorrhage, leading to high morbidity and mortality rates [2]. The most common form of severe leptospirosis is known as Weil's disease, characterized by jaundice, renal failure, and haemorrhage [3]. The most serious and life-threatening form of leptospirosis complications is known as severe pulmonary haemorrhage syndrome. Weil's disease and severe pulmonary haemorrhage syndrome cause death rates greater than 10% and 50% respectively even under treatment [4,5]. In addition, the most effective and safe antibiotics treatment for leptospirosis is yet to be found.
Leptospirosis poses a diagnostic challenge for physicians because of its variable severity. The diagnosis of severe leptospirosis through clinical assays such as traditional culture, serologic tests and pathogen-specific PCR assays is difficult due to unsatisfactory sensitivity and specificity [6]. The development of nextgeneration sequencing technique has revolutionized our ability to discover emerging pathogens, especially new viruses [7]. However, the clinical applications of next-generation sequencing for the diagnosis of infectious diseases are still to be explored [8]. We here report a case of severe leptospirosis with accurate and quick diagnosis by unbiased next-generation sequencing in order to administer appropriate treatment.
Case Presentation
A 35-year-old male presented to another hospital with fever, jaundice, yellow urine, oligouria on July 29, 2014 (Figure 1A). The patient had been in Hunan province, China, in July 5, 2014, and began to have fever after his coming back to Shenzhen in July 26, 2014. Then he complained diffuse weakness, decreased appetites, fear of cold, headache, backache, unable to walk caused by anchylosis and heavy ache in both legs in the following two days (Figure 1A). He was previously healthy and the only risk factor for the specific infection was household exposure in Hunan (Figure 1A).
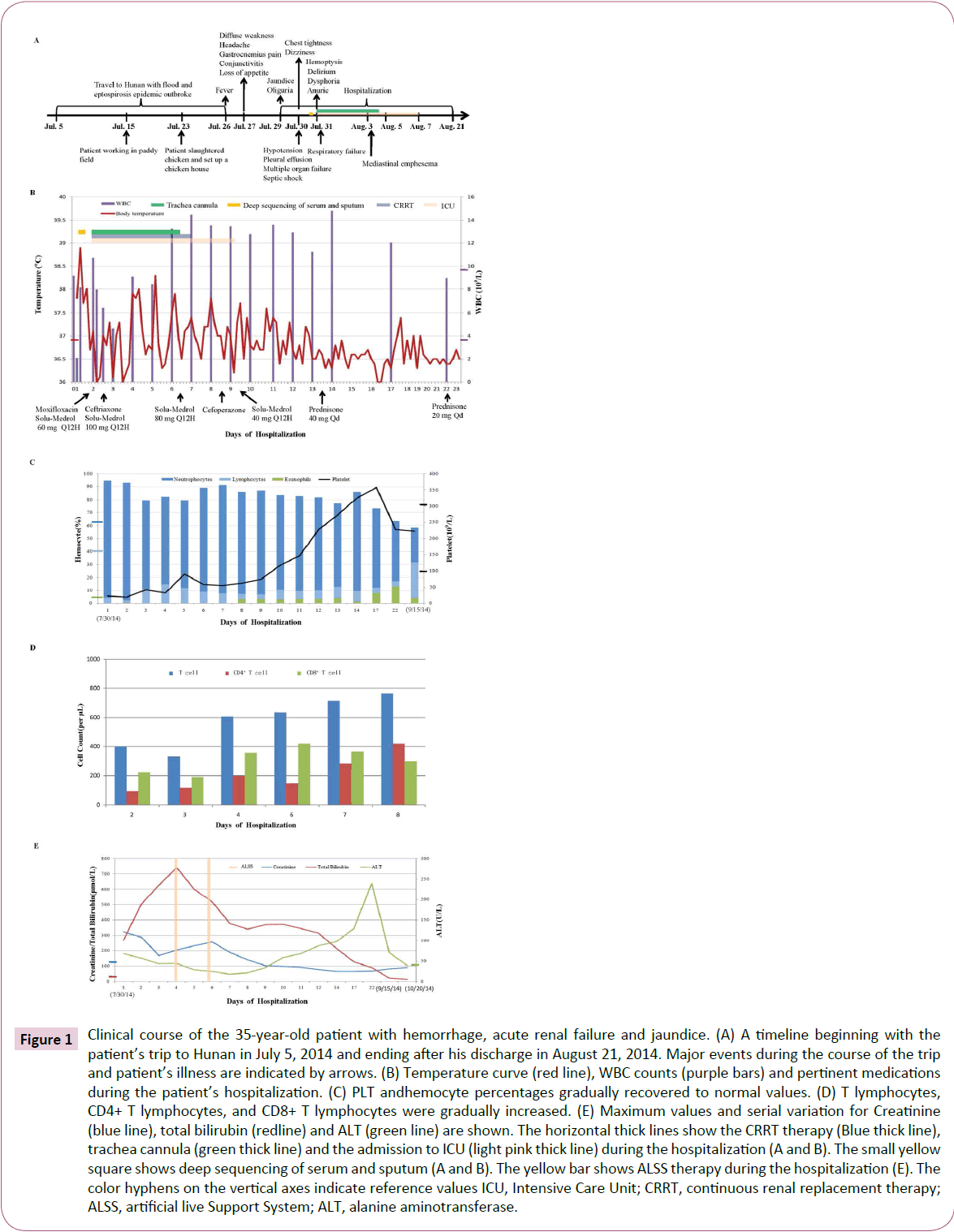
Figure 1: Clinical course of the 35-year-old patient with hemorrhage, acute renal failure and jaundice. (A) A timeline beginning with the patient’s trip to Hunan in July 5, 2014 and ending after his discharge in August 21, 2014. Major events during the course of the trip and patient’s illness are indicated by arrows. (B) Temperature curve (red line), WBC counts (purple bars) and pertinent medications during the patient’s hospitalization. (C) PLT andhemocyte percentages gradually recovered to normal values. (D) T lymphocytes, CD4+ T lymphocytes, and CD8+ T lymphocytes were gradually increased. (E) Maximum values and serial variation for Creatinine (blue line), total bilirubin (redline) and ALT (green line) are shown. The horizontal thick lines show the CRRT therapy (Blue thick line), trachea cannula (green thick line) and the admission to ICU (light pink thick line) during the hospitalization (A and B). The small yellow square shows deep sequencing of serum and sputum (A and B). The yellow bar shows ALSS therapy during the hospitalization (E). The color hyphens on the vertical axes indicate reference values ICU, Intensive Care Unit; CRRT, continuous renal replacement therapy; ALSS, artificial live Support System; ALT, alanine aminotransferase.
Initial blood tests revealed total white blood cell (WBC) count 9.18 × 109/L with neutrophils percentage 89.8% and platelet (PLT) count 14 × 109/μl. Serum creatinine (Cr) was 146 μmol/L and serum urea nitrogen (BUN) was 8.5 mmol/L. Total bilirubin (TB) was 113 μmol/L, lactate dehydrogenase (LDH) 293 U/L, and alanine transaminase (ALT) 68 IU/L, creatine kinase (CK) 3980 U/L, amylase (AMY) 273 U/L. Urine tests showed protein positive. Ultrasonography showed normal appearances of his , bile ducts, pancreas, but a slight enlargement of the spleen. He was injected with diammonium glycyrrhizinate, glutathione, acetylsalicylate, dexamethasone, platelet and plasma. In July 30, within 10 hrs of first admission he became hypotensive with blood pressure (BP) 60/40 mmHg, presented sweat, cold limbs, chest tightness and chest pain (Figure 1A). He remained hypotensive with BP 80/60 mmHg after receiving 0.5 L hydroxyethyl starch plus sodium chloride administration of intravenous fluid. Then he was transferred to the Infectious Diseases Unit in our hospital for further therapy and investigation.
On admission the patient showed severe icteric, gastrocnemius pain. He had normal vital signs except body temperature 37.8°C and unremarkable physical examination except moist rale in both lungs. He was highly suspicious with leptospirosis and epidemic hemorrhagic fever (EHF). However, both blood cultures and the rapid agglutination test for Leptospira were negative. Further immunoglobulin M (IgM) and IgG antibody tests for Leptospira were also negative (by BGI, Shenzhen). Finally, through nextgeneration sequencing (by BGI, Shenzhen), we successfully identified Leptospira interrogans serovar Copenhageni from both the sputum and blood samples (Table S1-S3, Figure 2A-2C).
| Species |
Blood |
Sputum |
| Human |
32,533,182 |
26,253,702 |
| Fungi |
483 |
37 |
| Bacteria |
4227 |
417 |
| Viruses |
980 |
2546 |
| Others |
2,920,770 |
3,319,348 |
| Total reads |
35,460,622 |
29,578,596 |
The next-generation sequencing run of two samples, including a blood sample and a sputum sample obtained from this patient, yielded 35,460,622 and 29,578,596 raw reads, respectively. After human host sequences subtracted and reads aligned to microbe references, for blood sample, the vast majority reads came from bacterial genomes, and overwhelming majority reads of sputum sample belonged to viral genomes.
Table S1: Summary counts of next generation sequencing (NGS) reads.
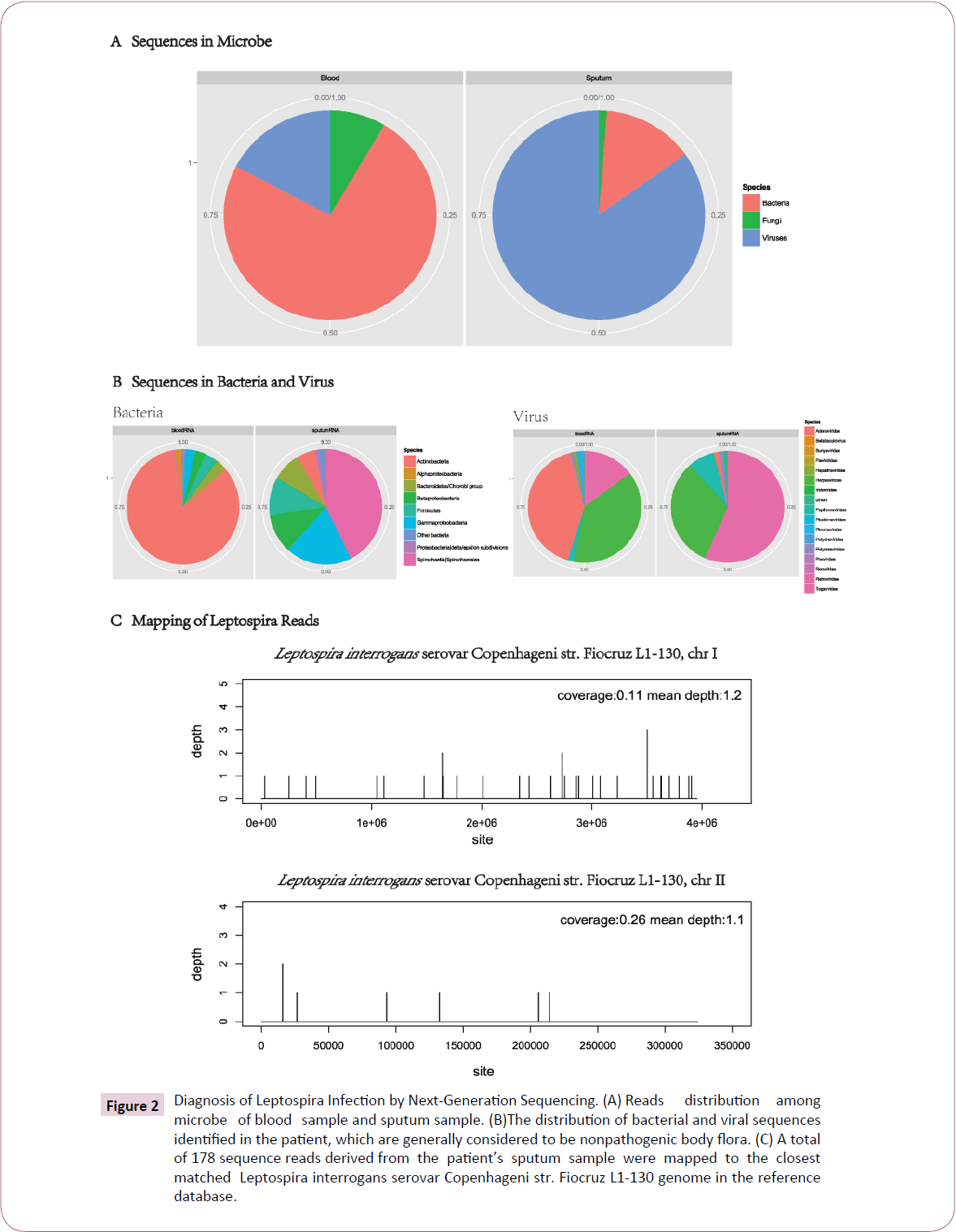
Figure 2: Diagnosis of Leptospira Infection by Next-Generation Sequencing. (A) Reads distribution among microbe of blood sample and sputum sample. (B)The distribution of bacterial and viral sequences identified in the patient, which are generally considered to be nonpathogenic body flora. (C) A total of 178 sequence reads derived from the patient’s sputum sample were mapped to the closest matched Leptospira interrogans serovar Copenhageni str. Fiocruz L1-130 genome in the reference database.
The next-generation sequencing run of two samples, including a sputum sample and a blood sample obtained from this patient, yielded 29,578,596 and 35,460,622 raw reads, respectively (Table S1 in the supplemental material). After subtraction of human host sequences the non-human reads were aligned to microbe reference sequences. There were 4227 (0.0119% of total) nonhuman reads (out of totally 2,926,460) belonged to bacterial genomes in the blood sample, whereas only 417 (0.0014% of total) reads of the sputum sample (3,322,348 non-human reads) belonged to bacterial genomes (Table S1 and Figure 2A). There were 980 (0.0028% of total) and 2,546 (0.0097% of total) reads mapped to virus sequences in the blood and sputum samples, respectively (Table S1 and Figure 2A). In bacterial reads of the sputum sample, 42.7% (178/417) (Table S2) corresponded to the Leptospiraceae family (Table S2 and Figure 2B). The mapped reads distributed along the Leptospira interrogans serovar Copenhageni str. Fiocruz L1-130 (NCBI accession number: NC_005823, NC_005824.1) with coverages of 0.11% and 0.26% for chromosomes 1 and 2, respectively (Figure 2C). In the blood sample, by contrast, 84% of bacterial reads (3549/4227) belonged to the Actinobacteria Phylum and there were only 25 reads mapped to the Leptospira genome (Table S2 and Figure 2B). Besides, we identified 980 and 2546 virus reads from blood sample and sputum sample, respectively. In the blood sample, the dominating virus corresponded to the family Adenoviridae (402 out of 980 reads; 41% of viral reads, ~ 0.01% of non-human reads and ~ 0.001% of total reads), and in the sputum sample, 56% (1438/2546) of the viral reads (0.04% of non-human reads and ~ 0.01% of total reads) belonged to the family of Retroviridae (Table S3, Figure2B). The patient was then supported with oxygen administration, treated initially with moxifloxacin, ademetionine, globulin, platelet and plasma (Figure 1B).
| Bacteria species categorization |
Blood RNA |
Sputum RNA |
| Cellular organisms/ Bacteria/ Spirochaetes/ Spirochaetia/ Spirochaetales |
25 |
178 |
| Cellular organisms/ Bacteria/ Cyanobacteria/ Nostocales/ Nostocaceae |
0 |
2 |
| Cellular organisms/ Bacteria/ Proteobacteria/ Betaproteobacteria/ Burkholderiales |
77 |
8 |
| Cellular organisms/ Bacteria/ Proteobacteria/ Alphaproteobacteria/ Caulobacterales |
3 |
0 |
| Cellular organisms/ Bacteria/ Actinobacteria/ Actinobacteria/ Actinobacteridae |
3549 |
21 |
| Cellular organisms/ Bacteria/ Bacteroidetes/ Chlorobi group/ Bacteroidetes/ Flavobacteriia |
5 |
11 |
| Cellular organisms/ Bacteria/ Firmicutes/ Clostridia/ Clostridiales |
100 |
27 |
| Cellular organisms/ Bacteria/ Bacteroidetes/ Chlorobi group/ Bacteroidetes/ Sphingobacteriia |
3 |
0 |
| Cellular organisms/ Bacteria/ Proteobacteria/ delta/ epsilon subdivisions/ Epsilonproteobacteria |
10 |
5 |
| Cellular organisms/ Bacteria/ Proteobacteria/ Betaproteobacteria/ Unclassified Betaproteobacte |
4 |
1 |
| Cellular organisms/ Bacteria/ Cyanobacteria/ Oscillatoriales/ Leptolyngbya |
1 |
0 |
| Cellular organisms/ Bacteria/ Cyanobacteria/ Pleurocapsales/ Chroococcidiopsis |
1 |
0 |
| Cellular organisms/ Bacteria/ Proteobacteria/ Gammaproteobacteria/ Pasteurellales |
0 |
20 |
| Cellular organisms/ Bacteria/ Firmicutes/ Clostridia/ Thermoanaerobacterales |
0 |
2 |
| Cellular organisms/ Bacteria/ Proteobacteria/ Betaproteobacteria/ Neisseriales |
39 |
36 |
| Cellular organisms/ Bacteria/ Chrysiogenetes/ Chrysiogenetes/ Chrysiogenales |
1 |
0 |
| Cellular organisms/ Bacteria/ Cyanobacteria/ Oscillatoriales/ Microcoleus |
3 |
0 |
| Cellular organisms/ Bacteria/ Thermotogae/ Thermotogae/ Thermotogales |
1 |
0 |
| Cellular organisms/ Bacteria/ Firmicutes/ Bacilli/ Lactobacillales |
17 |
8 |
| Cellular organisms/ Bacteria/ Chlamydiae/ Verrucomicrobia group/ Verrucomicrobia/ Opitutae |
1 |
0 |
| Cellular organisms/ Bacteria/ Proteobacteria/ Gammaproteobacteria/ Enterobacteriales |
30 |
31 |
| Cellular organisms/ Bacteria/ Deinococcus-Thermus/ Deinococci/ Deinococcales |
3 |
0 |
| Cellular organisms/ Bacteria/ Proteobacteria/ Alphaproteobacteria/ Sphingomonadales |
12 |
0 |
| Cellular organisms/ Bacteria/ Elusimicrobia/ environmental samples/ Uncultured Termite group |
0 |
1 |
| Cellular organisms/ Bacteria/ Proteobacteria/ Betaproteobacteria/ Methylophilales |
2 |
1 |
| Cellular organisms/ Bacteria/ Proteobacteria/ Betaproteobacteria/ Nitrosomonadales |
1 |
0 |
| Cellular organisms/ Bacteria/ Actinobacteria/ Actinobacteria/ Coriobacteridae |
0 |
1 |
| Cellular organisms/ Bacteria/ Proteobacteria/ Gammaproteobacteria/ Methylococcales |
1 |
0 |
| Cellular organisms/ Bacteria/ Proteobacteria/ Betaproteobacteria/ Rhodocyclales |
2 |
0 |
| Cellular organisms/ Bacteria/ Proteobacteria/ Gammaproteobacteria/ Xanthomonadales |
12 |
1 |
| Cellular organisms/ Bacteria/ Unclassified Bacteria/ Unclassified Bacteria (miscellaneous)/ Bac |
1 |
0 |
| Cellular organisms/ Bacteria/ Bacteroidetes/ Chlorobi group/ Bacteroidetes/ Cytophagia |
2 |
0 |
| Cellular organisms/ Bacteria/ Proteobacteria/ Alphaproteobacteria/ Rhizobiales |
66 |
0 |
| Cellular organisms/ Bacteria/ Firmicutes/ Negativicutes/ Selenomonadales |
39 |
3 |
| Cellular organisms/ Bacteria/ Tenericutes/ Mollicutes/ Mycoplasmatales |
1 |
4 |
| Cellular organisms/ Bacteria/ Bacteroidetes/ Chlorobi group/ Bacteroidetes/ Bacteroidia |
122 |
23 |
| Cellular organisms/ Bacteria/ Deferribacteres/ Deferribacteres/ Deferribacterales |
1 |
0 |
| Cellular organisms/ Bacteria/ Dictyoglomi/ Dictyoglomia/ Dictyoglomales |
1 |
0 |
| Cellular organisms/ Bacteria/ Proteobacteria/ delta/ epsilon subdivisions/ Deltaproteobacteria |
2 |
0 |
| Cellular organisms/ Bacteria/ Firmicutes/ Bacilli/ Bacillales |
5 |
5 |
| Cellular organisms/ Bacteria/ Proteobacteria/ Gammaproteobacteria/ Pseudomonadales |
84 |
27 |
| Cellular organisms/ Bacteria/ Chloroflexi/ Caldilineae/ Caldilineales |
0 |
1 |
In the sputum sample, the majority of bacterial reads (178 of 417 reads; 42.7%) matched Leptospira genome of Leptospiraceae family referring to NCBI nucleotide reference database. In the blood sample, by contrast, most of bacterial reads (3549 of 4227; 84%) matched Actinobacteria family and only 25 reads belonged to Leptospira.
Table S2: Summary counts of bacterial reads.
| |
|
|
| Virus species categorization |
Blood RNA |
SputumRNA |
| Viruses|dsDNA viruses, no RNA stage|Papillomaviridae|Alphapapillomavirus |
17 |
185 |
| Viruses|dsRNAviruses|Reoviridae|Sedoreovirinae |
2 |
0 |
| Viruses|ssRNAviruses|ssRNA positive-strand viruses, no DNA stage|Flaviviridae |
0 |
1 |
| Viruses|Retro-transcribingviruses|Retroviridae|Orthoretrovirinae |
7 |
67 |
| Viruses|ssRNAviruses|ssRNA positive-strand viruses, no DNA stage|Togaviridae |
1 |
0 |
| Viruses|dsRNAviruses|Picobirnaviridae|Picobirnavirus |
2 |
0 |
| Viruses|dsDNA viruses, no RNA stage|Polydnaviridae|Bracovirus |
0 |
1 |
| Viruses|Retro-transcribingviruses|Hepadnaviridae|Orthohepadnavirus |
9 |
0 |
| Viruses|dsDNA viruses, no RNA stage|Poxviridae|Chordopoxvirinae |
1 |
2 |
| Viruses|dsDNA viruses, no RNA stage|Adenoviridae|Mastadenovirus |
402 |
35 |
| Viruses|dsDNA viruses, no RNA stage|Iridoviridae|unclassifiedIridoviridae |
0 |
1 |
| Viruses|dsDNA viruses, no RNA stage|Polyomaviridae|Polyomavirus |
4 |
30 |
| Viruses|dsDNA viruses, no RNA stage|Herpesvirales|Herpesviridae |
375 |
816 |
| Viruses|dsDNA viruses, no RNA stage|Baculoviridae|Betabaculovirus |
1 |
0 |
| Viruses|ssRNAviruses|ssRNA positive-strand viruses, no DNA stage|Picornavirales |
18 |
0 |
| Viruses|Retro-transcribingviruses|Retroviridae|unclassifiedRetroviridae |
138 |
1371 |
| Viruses|ssRNAviruses|ssRNA negative-strand viruses|Bunyaviridae |
1 |
1 |
| Viruses|dsDNA viruses, no RNA stage|Papillomaviridae|unclassifiedPapillomavirida |
0 |
7 |
| others |
2 |
29 |
Totally, 980 and 2546 virus reads from blood sample and sputum sample were identified respectively. For blood sample, the dominating virus matched Adenoviridae family (402 of 980 reads; 41%). In sputum sample, however, the reads of Retroviridae family (1438/ 2546) is the largest group.
Table S3: Summary counts of viral reads.
Within 3 hours of admission, his temperature rose to 38.9°C. He complained headache and gastrocnemius pain, chest tightness and dizziness (Figure 1A). His blood pressure was decreased to 78/42 mmHg, heart rate was 120 beats per minute. His oxygen saturation was 97% (FiO2: 33%), whereas the laboratory tests revealed a severe thrombocytopenia (22 × 109/μl), decreased WBC count (2.09 × 109/L) with marked elevation of neutrophils (94.7%). Coagulation parameters showed hyperfibrinolysis but prothrombin time activity (PTA) was normal (94%). His albumin was 20 g/L, hemoglobin 109 g/L, C-reactive protein level 90 mg/L. However, he also had an acute renal and liver failure (Cr levels: 323.5 μmol/L, ALT levels: 67.7 U/L, TB levels: 269.9 μmol/L), cardiorespiratory failure (troponin I levels: 0.145 ng/ ml, N-terminal pro-brain natriuretic peptide levels: 7320 pg/ ml, PaO2 levels: 69%) as well as elevated creatine kinase (1600 U/L), myohemoglobin (2000 ng/ml) and amylase (318.1 U/L). Arterial blood gas (ABG) analysis revealed type I respiratory failure, hypokalemia and metabolic acidosis. Urine examination showed severe albuminuria, leukocyturia and erythrocyturia. Electrocardiogram revealed segment ST and T abnormalities.
Initial chest radiograph (X-ray) showed pleural effusion on the right side (Figure 3A). Considering that the patient might be suffering from multiple organ failure (MOF) and septic shock, he was administrated intravenous methylprednisolone 60 mg every 12 hours (Q12H) (Figure 1B). However, he remained hypotensive despite receiving 1000 ml of intravenous fluid and 50 ml of albumin. Subsequently, his blood pressure was approximately normal (BP 96/59 mmHg) after required circulatory support with intravenous dopamine 180 mg pumping.
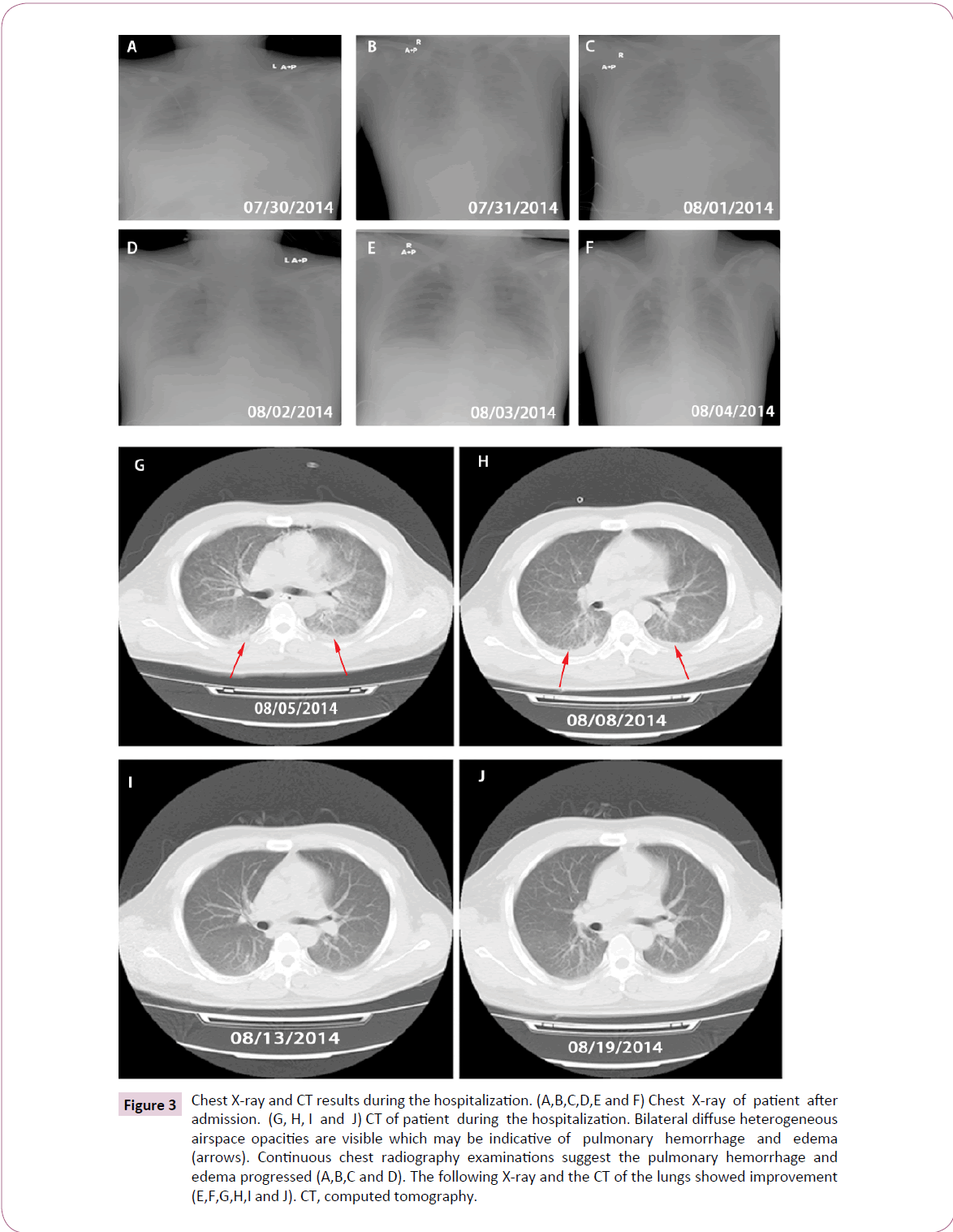
Figure 3: Chest X-ray and CT results during the hospitalization. (A,B,C,D,E and F) Chest X-ray of patient after admission. (G, H, I and J) CT of patient during the hospitalization. Bilateral diffuse heterogeneous airspace opacities are visible which may be indicative of pulmonary hemorrhage and edema (arrows). Continuous chest radiography examinations suggest the pulmonary hemorrhage and edema progressed (A,B,C and D). The following X-ray and the CT of the lungs showed improvement (E,F,G,H,I and J). CT, computed tomography.
During the following 9 hours, he began to cough, with gradually increasing chest pain, anhelation (breathing rate: 30-36 per minute), thrombocytopenia (19 × 109/μl), elevated WBC count (10.74 × 109/L) and neutrophils (93.3%). Despite circulatory support, he became anuric and continuous oxygen saturation decrease was initiated. Finally, he started to have hemoptysis and his respiratory function quickly deteriorated, necessitating supplies of higher concentration of oxygen for inhalation. He was given intravenous pituitrin followed by intravenous methylprednisolone and batroxobin was added. Unfortunately, he remained repeated hemoptysis, with delirium and dysphoria (Figure 1A). He presented with an unstable oxygen saturation ranged from 50% to 75% and was admitted to the Intensive Care Unit (ICU), where he underwent trachea cannula procedures, and the continuous renal replacement therapy (CRRT) was initiated (Figure 1A). A second chest X-ray showed signs suggestive of diffuse alveolar edema and intrapulmonary bleeding (Figure 3B). He was given intravenous methylprednisolone up to 100 mg, moxifloxacin replaced by Ceftriaxone, and norepinephrine 0.4 μg/kg/hour added (Figure 1B).
After trachea cannula, his respiratory function dramatically improved with stable oxygen saturation up to 94%. His highest temperature gradually decreased although still suffered from repeated fever. Blood tests revealed normal WBC count but still with marked elevation of neutrophils (Figure 1B and 1C). His PLT and total T lymphocytes, CD4+ T lymphocytes, CD8+ T lymphocytes increased gradually in the following days (Figure 1C and 1D). Continuous chest radiography examinations revealed that the pulmonary abnormalities progressed (Figures 3A-3D). But three days later, the X-ray and the following computed tomography (CT) of the lungs showed improvement (Figure 3E-3J). After his stabilization he had the tracheal extubation in August 4 and his oxygen saturation remained 98% after the procedure. However, he showed elevated WBC count (13.3 × 109/L) again soon, and neutrophil count was still high (89.1%), cefoperazone plus sulbactam (2000 mg every 6 hours) were administered (Figure 1B). Details of the blood routine, timing of the use of antibiotics and hormones, as well as CRRT therapy and respiratory support during the hospitalization were shown in (Figure 1B and 1C). In August 7, he was transferred to the Infectious Diseases Unit for further investigation.
During the hospitalization, the patient also developed marked hyperbilirubinemia with TB 741.2 μmol/L and enzyme-jaundice separation occurred (Figure 1E). Two days after admission, the CRRT was initiated. TB and Cr levels remained increasing, but decreased after ALSS was used twice (Figure 1E). ALT level was decreasing except for a short elevation at the end. The patient gradually recovered after transferred to Infectious Diseases Unit. Finally, 3 weeks after hospitalization the patient was fully recovered and the follow-up examinations were normal with no further complications.
Discussion
Early diagnosis of leptospirosis is important otherwise effective treatment would not be carried out to achieve successful outcome [9]. However, diagnosis of leptospirosis is difficult not only because of the clinical complications but also the lack of powerful diagnostic techniques for laboratory use.
The standard diagnostic assay, serological testing like microscopic agglutination test (MAT), is the reference method but only reliable within one week after disease onset. Also, enzyme-linked immunosorbent assay (ELISA), traditional culture and polymerase chain reaction (PCR) detection methods require similar time span and has limited sensitivity given the variety of Leptospira species and lacking of consensus among different assays [2,10-12]. Moreover, these tests are usually negative when people have already taken antibiotics or under mild symptoms. In tropical regions, leptospirosis is often indistinguishable from fevers (like dengue) due to the poor availability of laboratory diagnostic tools, which largely impedes effective disease control [2,13]. Therefore, early diagnosis of leptospirosis with clarification of particular species is of vital importance, especially in Leptospira interrogans infected patients whose acute respiratory distress can progress very rapidly [11]. Importantly, the severity of clinical symptoms in Chinese leptospirosis patients seems to have a certain correlation with the different serovars of Leptospira interrogans [4].
In this case report, unbiased next-generation sequencing was applied for blood and sputum samples to generate a large number of reads spanning the genome of Leptospira interrogans serovar Copenhageni which belongs to the serogroup icterohaemorrhagiae. The sputum sample was more successful than the blood sample for providing convincing information on the presence of Leptospira. Our diagnosis of this patient with Leptospira interrogans infection is consistent with the geographic distribution (south china), risk factors (flood, paddy filed), timing (summer) and severe symptoms (Weil’s disease, pulmonary hemorrhage). The rapid and accurate diagnosis by unbiased next-generation sequencing is therefore very important for our effective treatment. This successful case implies that the nextgeneration sequencing technology has great application potential for determining the infectious agents in clinical samples [6].
Currently, immediate antibiotics treatment for suspected leptospirosis is controversial although the WHO recommends initiating treatment within five days of suspected leptospirosis onset [14,15]. Traditional antibiotics treatment includes the combination of penicillin, ampicillin, amoxicillin, doxycycline and even cefotaxime or ceftriaxone for more severe cases [16]. However, traditional interventions are not effective and safe enough for severe leptospirosis cases [2,17]. Our patient experienced a life-threatening illness with multiple complications potentially associated with severe leptospirosis. The patient’s condition further deteriorated and progressed to multiple organ dysfunctions. Intravenous Moxifloxacin was replaced by ceftriaxone, and then changed to cefoperazone for preventing further infection.
As reported in the literature, patients would benefit from antibiotics treatment combined with immunosuppressive agents, like corticosteroids, as immune mechanisms might be crucial to presumed pulmonary hemorrhages in advanced leptospirosis [18,19]. It has been observed that corticosteroids reduce mortality and change outcome significantly when used early in the management of pulmonary leptospirosis [19,20]. A systematic review indicates that four out of five studies demonstrated the benefit of corticosteroids in treating severe disease with pulmonary involvement when administered early in the course of the disease, only the randomized controlled trial study showed that corticosteroids are ineffective and may increase the risk of nosocomial infections [21]. Currently, there is no consensus on the corticosteroids treatment for severe leptospirosis with pulmonary involvement, as convincing evidence is still lacking [22]. In our case, methylprednisolone was used soon after his hospitalization and the dose was increased rapidly according to his responses, which may be a crucial reason for the favorable outcomes.
Conclusion
In summary, the application of unbiased next-generation sequencing provided a clinically actionable fast diagnosis for Leptospira infection and even for the identification of species and serovars. In addition, for leptospirosis patients with presumed pulmonary hemorrhage, corticosteroids and antibiotics combined administration in the early course of the disease is highly recommended. Our success provides valuable insight of next-generation sequencing application in not only Leptospira infection, but also other uncertain and unknown pathogens infection, so that prompt and effective clinical interventions would be prescribed.
Consent
Written informed consent was obtained from the patient for publication of this case report and any accompanying images. A copy of the written consent is available for review by the Editor of this journal.
Declarations
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article and its additional files.
Ethics approval and consent to participate and publish
Collection and analysis of the patient’s clinical samples in present study was approved by the ethics committee of Shenzhen Third People’s Hospital. Written informed consent was obtained from the patient for publication of this case report and any accompanying images. A copy of the written consent is available for review by the Editor of this journal.
Competing Interests
The authors declare that they have no competing interests.
Authors Contribution
Wen Zeng: Collected and analyzed data, manuscript writing; Liqiang Li: Project design, data analysis and interpretation, financial support, manuscript writing; Yuhai Bi: data analysis and interpretation, manuscript writing; Wei Fang: Data analysis and interpretation, manuscript writing; Jiansong Tang: data analysis and interpretation, manuscript writing; Jing Yuan: data analysis and interpretation; Jiandong Li: Performed experiments; Jianming Li: data analysis and interpretation; Rongrong Zou: Performed experiments and analyzed data; Na Pei: Data analysis; Huijue Jia: Financial support, data analysis and interpretation; Xin Liu: Financial support, data analysis and interpretation; Hui Wang: data analysis and interpretation, manuscript writing; Jinmin Ma: Conception and design, data analysis and interpretation, financial support, manuscript writing; Yingxia Liu: Project design, collection and/or assembly of data, data analysis and interpretation, financial support, manuscript writing and final approval of manuscript. All authors read and approved the final manuscript.
Acknowledgement
This work was supported by The Medical Key Discipline Project of Shenzhen (No.201161).
19248
References
- Costa F, Hagan JE, Calcagno J, Kane M, Torgerson P, et al. (2015) Global Morbidity and Mortality of Leptospirosis: A Systematic Review. PLoSNegl Trop Dis 9: e0003898.
- Helmerhorst HJ, Van Tol EN, Tuinman PR, De Vries PJ, Hartskeerl RA, et al. (2012) Severe pulmonary manifestation of leptospirosis. Neth J Med 70: 215-221.
- Ko AI, Goarant C, Picardeau M (2009) Leptospira: the dawn of the molecular genetics era for an emerging zoonotic pathogen. Nature Reviews Microbiology 7: 736-747.
- Hu W, Lin X, Yan J (2014) Leptospira and leptospirosis in China. CurrOpin Infect Dis 27: 432-436.
- Mcbride AJA, Athanazio DA, Reis MG, Ko AI (2005) Leptospirosis. Current Opinion in Infectious Diseases 18: 376-386.
- Wilson MR, Naccache SN, Samayoa E, Biagtan M, Bashir H, et al. (2014) Actionable diagnosis of neuroleptospirosis by next-generation sequencing. N Engl J Med 370: 2408-2417.
- Deurenberg RH, Bathoorn E, Chlebowicz MA, Couto N, Ferdous M, et al. (2017) Application of next generation sequencing in clinical microbiology and infection prevention. J Biotechnol 243: 16-24.
- Dunne WM, Westblade LF, Ford B (2012) Next-generation and whole-genome sequencing in the diagnostic clinical microbiology laboratory. Eur J ClinMicrobiol Infect Dis 8: 1719-1726.
- Organization WH (2011) Report of the second meeting of the Leptospirosis Burden Epidemiology. Reference Group 30.
- Taylor D, Karamadoukis L (2013) Plasma exchange in severe leptospirosis with multi-organ failure: a case report. J Med Case Rep 7: 169.
- Lau CL, Depasquale JM (2012) Leptospirosis, diagnostic challenges, American Samoa. Emerg Infect Dis 18: 2079-2081.
- Bourhy P, Bremont S, Zinini F, Giry C, Picardeau M (2011) Comparison of real-time PCR assays for detection of pathogenic Leptospira spp. in blood and identification of variations in target sequences. J ClinMicrobiol 49: 2154-2160.
- Blacksell SD, Newton PN, Bell D, Kelley J, Mammen MP, et al. (2006) The comparative accuracy of 8 commercial rapid immunochromatographic assays for the diagnosis of acute dengue virus infection. Clinical Infectious Diseases 42: 1127-1134.
- Charan J, Saxena D, Mulla S, Yadav P (2013) Antibiotics for the treatment of leptospirosis: systematic review and meta-analysis of controlled trials. Int J Prev Med 4: 501-510.
- Organization WH (2003) Human leptospirosis: guidance for diagnosis, surveillance and control 125.
- Isa SE, Onyedibe KI, Okolo MO, Abiba AE, Mafuka JS, et al. (2014) A 21-Year-Old Student with Fever and Profound Jaundice. Plos Neglected Tropical Diseases 8: e2534.
- Brett-Major DM, Coldren R (2012) Antibiotics for leptospirosis. Cochrane Database Syst Rev 2: CD008264.
- Schulze MH, Raschel H, Langen HJ, Stich A, Tappe D (2014) Severe LeptospirainterrogansserovarIcterohaemorrhagiae infection with hepato-renal-pulmonary involvement treated with corticosteroids. Clin Case Rep 2: 191-6.
- Jayakrishnan B, Ben Abid F, Balkhair A, Alkaabi JK, Al-Rawas OA, et al. (2013) Severe Pulmonary Involvement in Leptospirosis: Alternate antibiotics and systemic steroids. Sultan QaboosUniv Med J 13: 318-322.
- Shenoy VV, Nagar VS, Chowdhury AA, Bhalgat PS, Juvale NI (2006) Pulmonary leptospirosis: an excellent response to bolus methylprednisolone. Postgrad Med J 82: 602-606.
- Rodrigo C, Lakshitha De Silva N, Goonaratne R, Samarasekara K, et al. (2014) High dose corticosteroids in severe leptospirosis: a systematic review. Trans R Soc Trop Med Hyg 108: 743-750.
- Hingorani RV, Kumar R, Hegde AV, Soman RN, Sirsat RA, et al. (2016) Is it Time to Rethink the Use of Steroids for Pulmonary Leptospirosis? J Assoc Physicians India 64: 78-79.








