Keywords
Dormancy, Developmental biology, Dechorionation, Dessication, Nuclei staining, Hoechst dye
Introduction
Rotifer background
The Rotifera phylum consist of approximately 2000 species of relatively small, pseudocoelomate, aquatic invertebrate that predominately populate freshwater lakes and ponds, and can contribute up to 30% of the total plankton biomass [1]. Brachionus plicatilis Müller (Rotifera, Monogononta) can serve as interesting model organism for studying the developmental biology in nonsegmented animals [2] owing to a short developmental cycle of ~20 hours [3]. In addition, B. plicatilis can be used to study the mechanism for primordial germ cells (PGCs) differentiation sharing conserved germ-line markers such as vasa and nanos [4]. Although Bdelloid rotifers can enter dormancy, by either whole body dessication or the formation of long-term surviving eggs; the monogonont rotifers enter dormancy during the egg stage either as the product of asexual parthenogenesis or by producing encysted diapausing resting egg (RE) formed from a fertilized ova [5].
Genetic screening of genes involved in dormancy
Initial screening of expressed sequence tags (EST) of B. plicatilis cultures in differing stages of development revealed the presence of genes associated with protection against reactive oxygen species (ROS), chaperone proteins (heat shock proteins), late embryogenesis abundant (LEA) proteins, trehalose metabolism, aquaporins and others in resting eggs [6]. Real time RT-PCR showed that LEA proteins and small heat shock proteins (sHSP) were upregulated in both resting eggs and females producing resting eggs in comparison to amictic eggs and females producing amictic eggs, respectively [6]. High-throughput Illumina sequencing data in concurrence to RT-PCR analysis showed higher expression of LEAs, glutathione-S-trasnferases, HSPs, and downregulation in trehalose metabolism, aquaporins transcription during early RE dormancy and the respective expression pattern for the aforementioned genes in females carrying REs versus amictic correlated with the types of eggs they produced [7]. Highthroughput Illumina sequencing was also used to show that for resting eggs, genes associated with cellular maintenance and survival were upregulated such as antioxidants, oxidoreductases, and protein chaperone genes; in comparison to amictic eggs, genes associated with cellular and organismic development such as cell cycle and cytoskeletal genes [8]. Differential transcription and expression for respective class 3 LEA genes and proteins have been shown to be temporary regulated during RE development [9]. The quizzical nature of LEA proteins may be explained by protein modeling algorithms that predict that native unfolded structure of LEA is transformed into folded proteins during desiccation to function in nuclear stabilization such as in DNA binding and cytosekeletal substitution [10].
Rotifer nuclei
Transmission electron microscopy (TEM) has identified several organelles including nuclei, mitochondria, and free ribosomes, in both the ectoderm and inner mass in resting eggs (RE) of Brachionus suspended in diapuase [11]. The nuclei of the ectodermal region lack nucleoli and have scattered mass of heterochromatin; whereas, the nuclei of the inner region contain nucleoli and condensed masses of chromatin [11]. In experiments by Hagiwara et al. [12], to determine hatching of B. plicatilis resting eggs in response to irradiation and other stimulants (e.x. hydrogen peroxide), noted that number of nuclei in resting eggs on day one after formation to day 6 increased from 22-39. The nuclei number for resting eggs in Brachionus plicatilis has been recorded to vary in the range between 26-160 [13]. Snell et al. [14] described a method for the decapsulation of rotifers for siRNA treatment, however this method was insufficient for the proper labeling by Hoechst staining of developing rotifers in our hands (data not shown). For this report, we describe a method to decapsulate encysted resting eggs for nuclei staining, which may ultimately be used in the determination of nuclei number in the development of resting eggs.
Methods
Maintenance of rotifers and specimen collection
B. plicatilis cultures were initiated at approximate density of 6 rotifers per 1 ml of 40 parts per thousand (ppt) sea water for a total volume of 500 ml in a 1 liter Erlenmeyer flask (~3,000 rotifers). The cultures were fed daily with 500 μl algae (~9 × 1011) and were grown logarithmically for 4-5 days at 25oC to achieve densities of between 40-50 rotifers per ml. B. plicatilis cultures were renewed every 7-10 days. Naïve young females were collected into round-bottom 96 well plates and were examined hourly for the presence of newly developed eggs. The timing for the bearing of new amicitic (AM) or resting eggs (RE) was recorded and 10-20 samples were separated and collected based on differing time points (Example: 6, 12, 18 hours after the eggs were visible outside the mother). The eggs were immediately transferred and stored at -80oC until they were used for staining experiments.
Decapsulation and Hoechst Staining
Decapsulation
For the samples that were decapsulated before subjected to staining, decapsulation was performed by the following procedure: Each set of resting eggs (10-20 eggs) was suspended in 400 μl in 40 ppt, 300 μl 40 ppt pH10, and 300 μl fresh 11% hypochlorite and vortexed for 4s (setting 7). Each solution containing REs was incubated for 3 minutes on ice. Next, the contents of the RE-containing solution was transferred into 5 ml 40 ppt and transferred into a 33 ml sieve. The sieve was washed with 5 ml of a 0.1% thiosulfate solution and the eggs were washed with PBS.
Staining
Amictic and resting eggs were stained using Hoechst dye (Sigma Aldrich-St. Louis, MO) by the following procedure: The eggs were briefly washed with PBS (5 minutes) and incubated for 1 hour in 4% para-formaldehyde resuspended in PBS and rewashed with PBS. Next the eggs were dehydrated by consecutive suspension in increasing ethanol concentrations (20%, 40%, 60%, 80%, diluted with PBS) for 10 min incubation in each solution. The eggs were stored overnight at 100% ethanol at -20°C. The following day, the eggs were rehydrated in decreasing ethanol concentrations (100%, 80%, 60%, 40%, 20%, diluted with PBS, and pure PBS) for 10 min incubation in each solution. The eggs were subsequently incubated in a PBS solution supplemented with 1% Triton X-100 for 5 min at room temperature. Followed by 30 minute incubation in a PBS solution supplemented with 3% Triton X-100 and 1% Tween-20. The eggs were stained with Hoechst solution at 1 μl/ ml PBS for 30 min at room temperature and briefly washed with PBS. For microscopic observation, the eggs were transferred into plates and covered with PBS.
Microscopy
A Leica Wide Field Microscope (Leica Model DMi8 inverted microscope) was used to take images of rotifer amictic and resting eggs collected at varying time point stained with Hoechst dye. After the images were captured, deconvulation software (Leica LASX software) was used to analyze the images and measure the number, size and intensity of observed nuclei.
Results
Maintenance of rotifers and specimen collection
B. plicatilis rotifers were continuously propagated under lab conditions in a simple salt solution fed with algae. For amictic eggs (Figure 1A-1C) and REs that were stained before the crucial 12 hour period (Figure 1D-1F), decapsulation was unnecessary. Both samples in Figure 1 were collected at 6 hours after the eggs were extruded from the mother. For the AM at 6 hours about 100 nuclei are observed as compared to the 50 observed in resting egg of 6 hours. After the 12 hour period, REs were decapsulated to observe and count the nuclei number (Figure 2). For REs collected at 12 hours (Figure 2A to 2F), about 50 nuclei are observed, as compared to the ~400 observed at the 18 hour period (Figure 2G and 2H). For this report, we optimized a decapsulation procedure in terms of hypochlorite strength, and bleaching duration for the nuclei labeling of encysted B. plicatilis resting eggs. During the decapsulation procedure, roughly 50% of the eggs are unusable for staining because of either overbleaching which can destroy the integrity of the egg, or under bleaching (Figure 2A and 2B) which does not allow for the Hoechst stain to penetrate the egg shell.
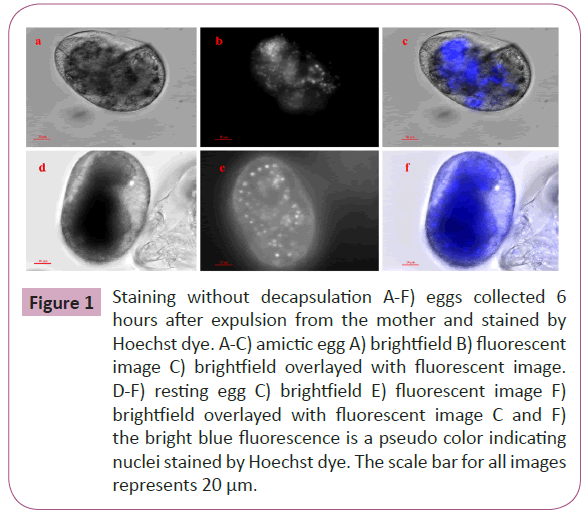
Figure 1: Staining without decapsulation A-F) eggs collected 6 hours after expulsion from the mother and stained by Hoechst dye. A-C) amictic egg A) brightfield B) fluorescent image C) brightfield overlayed with fluorescent image. D-F) resting egg C) brightfield E) fluorescent image F) brightfield overlayed with fluorescent image C and F) the bright blue fluorescence is a pseudo color indicating nuclei stained by Hoechst dye. The scale bar for all images represents 20 μm.
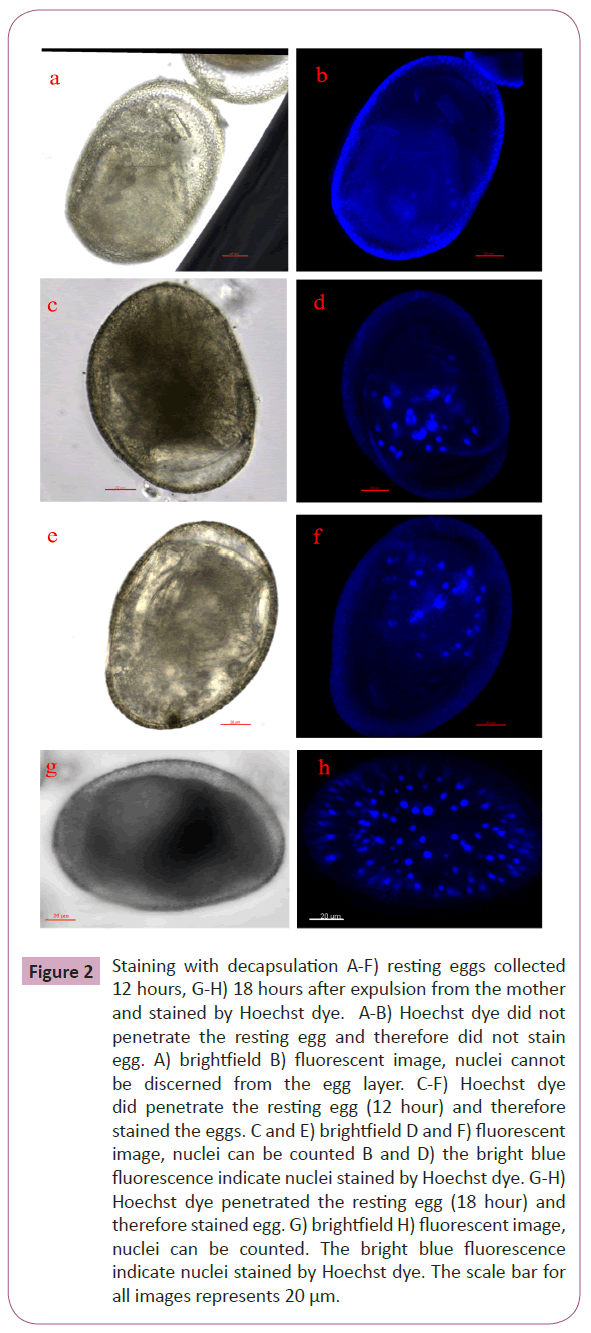
Figure 2: Staining with decapsulation A-F) resting eggs collected 12 hours, G-H) 18 hours after expulsion from the mother and stained by Hoechst dye. A-B) Hoechst dye did not penetrate the resting egg and therefore did not stain egg. A) brightfield B) fluorescent image, nuclei cannot be discerned from the egg layer. C-F) Hoechst dye did penetrate the resting egg (12 hour) and therefore stained the eggs. C and E) brightfield D and F) fluorescent image, nuclei can be counted B and D) the bright blue fluorescence indicate nuclei stained by Hoechst dye. G-H) Hoechst dye penetrated the resting egg (18 hour) and therefore stained egg. G) brightfield H) fluorescent image, nuclei can be counted. The bright blue fluorescence indicate nuclei stained by Hoechst dye. The scale bar for all images represents 20 μm.
Discussion
General dormancy
Dormancy is a phenomenon that encompasses a period of suspended metabolism and arrested development by intracellular and intranuclear factors [15]. Desiccation tolerance involves a serious of coordinated events involving late embryogenesis abundant (LEA) and heat shock proteins (HSPs) that are activated during water loss to prevent the drying out of cells [16-18]. In plants, pollen and seeds, survive desiccation by recruiting soluble non-reducing carbohydrate and LEAs to form intracellular glass that slow down intracellular trafficking and stabilize macromolecules [19]. Due to the unstructured conformation of LEA proteins, the classical role for these proteins is believed to function as a hydration buffer or in water substitution [20,10]. LEA proteins undergo conformational changes during anyhdyrobiotic and heat stress conditions that may function in cellular stabilization. The mitochondrial localization of a LEA has been reported in the brine shrimp, Artemia franciscana where it is expected to serve in a bioprotective role during anyhydrobiosis [21].
Dormancy genes
Genes for the trehalose biosynthetic pathway, which produces a product that is accumulated during temperature or osmotic stress, are ubiquitously found across the three forms of life and in kingdoms that include fungi, plants and animals [22]. Trehalose is a carbohydrate that is ubiquitously synthesized in a variety of lower and higher organisms to serve in a bioprotective role for protein degradation during oxidative stress and/or dehydration [23]. Trehalose has a role in water replacement during in both anyhdrobiotic and vitrificatic states associated with dormancy [24]. LEAs from Artemia franciscana have been shown to protect function in enzymes during desiccation and to work synergistically with trehalose to provide better protection than either agent acting in singular [25]. In addition, several small heat shock proteins, such as (sHSP) p26 and ArHsp22 have been has identified to localize to the cytoplasm and nucleus in Artemia and are thought to provide protection against irreversible stress incduced protein denaturation during encystment and diapause [15,26,27].
Molecular manipulation in rotifers
From the two possible modes of dsRNA uptake for rotifers that include injestion via feeding of young rotifers or during rehydration of dried resting eggs, initial trials showed higher intake by the former method [28]. Conjugated dsRNA to lipofectant have shown varying degrees of gene knockdown and phenotypic changes targeting genes for reproduction and lectin-binding in the rotifer Brachionus manjavacas [14,29]. For example, a gene that is involved in eye formation in higher organisms termed Pax6 was knocked-down by feeding rotifers E. coli exoressing dsRNA [30]. However, Snell et al. [14] showed that the RNAi pathway can also be activated by the addition of dsRNA to decapsulated rotifer resting eggs. In our hands, the suggested protocol of using 6% hypochlorite for 1 minute at room temperature did not dechorinate the eggs sufficiently for staining with Hoechst dye. In our modified procedure, we dechorionated the resting eggs by bleaching with 11% hypochlorite for 3 minutes on ice and rapidly washing out the bleach with sea water and neutralizing the hypochlorite with thiosulfate. For this method, approximately 50% of the eggs were properly decapsulated to allow for staining with Hoechst dye (Figure 2). As compared to amictic eggs, the division of nuclei occurs slower for the first 12 hours in resting eggs (data not shown).
Conclusion
Rotifers can serve as interesting model to study dormancy and developmental biology. In addition, the bdelloid rotifer (Macrotrachela quadricornifera) was proposed to be used in space experiments aboard the international space station (ISS) for understanding the role of cytoskeletal organization in the establishment of the division furrow and role in the embryonic developmental stages, including oogenesis in hypogravity conditions [31,32] and can be an effective agent in bioremediation [33]. Previous reports have shown that B. plicatilis is amenable to genetic manipulation by the RNAi pathway [28]. However, the methodology used to insert dsRNA into encysted resting eggs was not efficient in our hands to penetrate the REs for nuclei staining with Hoechst dye. Therefore, we adapted the Snell et al. protocol for decapsulation of REs to allow for proper staining of nuclei to determine nuclear progression during the development of fertilized resting eggs in Brachionus plicatilis Müller.
8799
References
- Lubzens E, Zmora O (2003) Production and nutritional value of rotifers. Live feeds in marine aquaculture 300-303.
- Boell LA, Bucher G (2008) Whole-mount in situ hybridization in the Rotifer Brachionusplicatilis representing a basal branch of lophotrochozoans. Development genes and evolution 218: 445-451.
- Paez, MC, Kurokura H, Kasahara H (1988) Embryonic development of amictic eggs of a rotifer Brachionusplicatilis. J Fac Appl Biol Sci 27: 93-99.
- Smith JM, Cridge AG, Dearden PK (2010) Germ cell specification and ovary structure in the rotifer Brachionusplicatilis. Evodevo 1: 5.
- Denekamp NY, Suga K, Hagiwara A, Reinhardt R, Lubzens E (2010) A role for molecular studies in unveiling the pathways for formation of rotifer resting eggs and their survival during dormancy. In Dormancy and Resistance in Harsh Environments 109-132.
- Denekamp NY, Thorne MA, Clark MS, Kube M, Reinhardt R, et al. (2009) Discovering genes associated with dormancy in the monogonont rotifer Brachionusplicatilis. BMC Genomics 10: 108.
- Denekamp NY, Reinhardt R, Albrecht MW, Drungowski M, Kube M, et al. (2011) The expression pattern of dormancy-associated genes in multiple life-history stages in the rotifer Brachionusplicatilis. Hydrobiologia 66: 51-63.
- Clark MS, Denekamp NY, Thorne MA, Reinhardt R, Drungowski M, et al. (2012) Long-term survival of hydrated resting eggs from Brachionusplicatilis. PLoS One 7: e29365.
- Denekamp NY, Reinhardt R, Kube M, Lubzens E (2010) Late embryogenesis abundant (LEA) proteins in nondesiccated, encysted, and diapausing embryos of rotifers. Biol Reprod 82: 714-724.
- Wise MJ, Tunnacliffe A (2004) POPP the question: what do LEA proteins do? Trends Plant Sci 9: 13-17.
- Wurdak ES, Gilbert JJ, Jagels R (1978) Fine structure of the resting eggs of the rotifers Brachionuscalyciflorus and Asplanchnasieboldi.Transactions of the American Microscopical Society 49-72.
- Hagiwara A, Hoshi N, Kawahara F, Tominaga K, Hirayama K (1995) Resting eggs of the marine rotifer Brachionusplicatilis Müller: development, and effect of irradiation on hatching. Hydrobiologia 31: 223-229.
- Boschetti C, Leasi F, Ricci C (2011) Developmental stages in diapausing eggs: an investigation across monogonont rotifer species. Hydrobiologia 66: 149-155.
- Snell TW, Shearer TL, Smith HA (2011) Exposure to dsRNA elicits RNA interference in Brachionusmanjavacas (Rotifera). Mar Biotechnol (NY) 13: 264-274.
- Willsie JK, Clegg JS (2001) Nuclear p26, a small heat shock/alpha-crystallin protein, and its relationship to stress resistance in Artemia franciscana embryos. J Exp Biol 204: 2339-2350.
- Hatanaka R, Hagiwara-Komoda Y, Furuki T, Kanamori Y, Fujita M, et al. (2013) An abundant LEA protein in the anhydrobiotic midge, PvLEA, acts as a molecular shield by limiting growth of aggregating protein particles. Insect biochemistry and molecular biology 1055-1067.
- Hoekstra FA, Golovina EA, Buitink J (2001) Mechanisms of plant desiccation tolerance. Trends Plant Sci 6: 431-438.
- Kikawada T, Nakahara Y, Kanamori Y, Iwata KI, Watanabe M, et al. (2006) Dehydration-induced expression of LEA proteins in an anhydrobioticchironomid. Biochemical and biophysical research communications 348: 56-61.
- Buitink J, Leprince O (2008) Intracellular glasses and seed survival in the dry state. C R Biol 331: 788-795.
- Hand SC, Menze MA, Toner M, Boswell L, Moore D (2011) LEA proteins during water stress: not just for plants anymore. Annu Rev Physiol 73: 115-134.
- Menze MA, Boswell L, Toner M, Hand SC (2009) Occurrence of mitochondria-targeted late embryogenesis abundant (LEA) gene in animals increases organelle resistance to water stress. Journal of Biological Chemistry 28: 10714-10719.
- Avonce N, Mendoza-Vargas A, Morett E, Iturriaga G (2006) Insights on the evolution of trehalose biosynthesis. BMC Evol Biol 6: 109.
- Jain NK, Roy I (2009) Effect of trehalose on protein structure. Protein Sci 18: 24-36.
- Sakurai M, Furuki T, Akao K, Tanaka D, Nakahara Y, et al. (2008) Vitrification is essential for anhydrobiosis in an African chironomid, Polypedilumvanderplanki. Proc Natl Acad Sci U S A 105: 5093-5098.
- Buitink J, Leprince O (2008) Intracellular glasses and seed survival in the dry state. C R Biol 331: 788-795.
- Qiu Z, Bossier P, Wang X, Bojikova-Fournier S, MacRae TH (2006) Diversity, structure, and expression of the gene for p26, a small heat shock protein from Artemia. Genomics 88: 230-240.
- Qiu Z, MacRae TH (2008) ArHsp2, a developmentally regulated small heat shock protein produced in diapause-destined Artemia embryos, is stress inducible in adults. FEBS journal 27: 3556-3566.
- Shearer TL, Snell TW (2007) Transfection of siRNA into Brachionusplicatilis (Rotifera). Hydrobiologia 59: 141-150.
- Snell TW, Shearer TL, Smith HA, Kubanek J, Gribble KE, et al. (2009) Genetic determinants of mate recognition in Brachionusmanjavacas (Rotifera). BMC Biol 7: 60.
- Dearden PK, Cridge A (2000) Expression and function of Pax genes in the rotifer Brachionusplicatilis. In Mech of development 12: 162-162.
- Ricci C, Boschetti C (2003) Bdelloid rotifers as model system to study developmental biology in space. Adv Space Biol Med 9: 25-39.
- Ricci C, Caprioli M, Boschetti C, Santo N (2005) Macrotrachelaquadricornifera featured in a space experiment. Hydrobiologia 53: 239-244.
- Kostopoulou V, CarmonaMJ, Divanach P (2012) The rotifer Brachionusplicatilis: an emerging bio-tool for numerous applications. Journal of biological research. Scientific annals of the school of biology, Aristotle University of Thessaloniki 17: 97-112.