Introduction
Aggregatibacter actinomycetemcomitans (Aa) is a capnophilic, Gram-negative coccobacillus, colonizing the human oral cavity [1,2]. Aa belongs to the normal oral flora in many healthy subjects, and is also implicated as a pathogen in certain aggressive forms of periodontal diseases such as the juvenile periodontitis [3]. A considerable genetic diversity and a variable ability to produce virulence factors have been found among Aa isolates [1,4].
Acquisition of Aa can start in early childhood and the rate of children colonized increases with their age [3]. The main source of acquisition is considered to be the close household contacts, such as between mothers carrying the pathogen and their children [5]. Early acquisition of potentially pathogenic oral bacterial species might impact the development of mucosal responses in the gingiva and may provide an enhanced risk for the development of periodontitis later in life [6]. Aa was found in periodontally healthy family members of Aa positive adults with periodontitis [7]. Based on serotyping, a possible Aa strain transmission has been suggested between parents and children with localized juvenile periodontitis [7] and between family members with aggressive periodontitis who were Aa-positive [8]. It is also well established that severe periodontitis clusters in families [9]. Nevertheless, about 33% of periodontally healthy children carrying Aa, had parents that did not possess the same pathogens [10]. Another study [11] found no Aa transmission between Brazilian women with severe chronic periodontitis and their children. Based on a literature review, [12] the authors concluded that transfer of bacteria between spouses occurs, but it appears to happen infrequently, however the transfer of organisms does not necessarily result in colonization or infection of the host. Furthermore, individuals who harbor putative pathogens frequently do not manifest any signs of periodontal disease
Seven distinct serotypes and several genotypes of Aa have been identified. Serotypes a, b, and c are the most predominant, while serotypes d, e, f, and g are more rarely isolated from oral samples [13-16].
The serotype association to the disease is rather unclear [17] and genetic factors, geographic origin, age and socioeconomic status seem to influence the serotype distribution [1,9]. Strains of serotype b are frequently found in subjects with localized juvenile periodontitis [18], while strains of serotype a appear more often in healthy subjects or subjects with mild forms of periodontal infection. Contradictory results are reported for serotype c strains that appear associated with both extra-oral infections and severe periodontitis and periodontal health [15].
Aa produces two exotoxins, a cytolethal distending toxin and a leukotoxin [1]. The former kills host cells by blocking their proliferation [19], while leukotoxin selectively destroys human cells of hematopoetic origin by disrupting the membrane integrity [20]. The production of leukotoxin greatly varies. Among the high toxin producers are strains that belong to a serotype b clone, the JP2 genotype, characterized by a deletion in the leucotoxin gene operon. Strains of JP2 genotype are particularly implicated in the pathogenesis of aggressive periodontitis [1,4].
Worldwide, the bacterium has been detected in oral samples of children and adolescents, irrespective of their periodontal health [21]. The prevalence found in these studies varied from 3% to 78%. In Greek children, the prevalence of Aa was previously examined in two studies; however, the reported results have been strongly conflicting; in a group of 40 healthy children, Aa was detected in 2.8% and 3.3% of children with permanent and primary dentition, respectively [21]. In the second study [22] that focused on the microbiota of various oral surfaces in 93 healthy children, Aa was found in all 93 children. In none of these studies, serotype or genotype identification of the strains was attempted.
The present study was conducted to examine the extent of Aa transmission from mothers to children and also to elucidate the Aa prevalence and the distribution of its serotypes in dental plaque of periodontally healthy Greek children and their mothers. Furthermore, the occurrence of strains from the highly leukotoxic clone JP2 was explored in this population.
Methods
Participants
A total of 108 children (58 boys and 50 girls) aged 5-12 years (Median age 100 months, min=45 max=162) and 83 mothers participated in the study. The children included were healthy (ASA I, II) and co-operative with no clinical and radiographical signs of periodontitis (no bone loss and deep pockets). The extent of dental plaque accumulation greatly varied among the children. They visited the clinics of Paediatric Dentistry in the Dental School of Aristotle University of Thessaloniki and that of Social Insurance Institute of Thessaloniki for oral check-up and eventually dental treatment during the period 2010-2011. All participants had no history of antibiotic therapy during the previous three months. The study was approved by the Ethics Committee of the Dental School of Aristotle University of Thessaloniki. Informed consent was obtained from all participants and their guardians.
Sample collection and microbiological analysis
Dental plaque was collected using a micro-brush from all buccal tooth surfaces. The plaque sample was transported in sterile potassium Phosphate-Buffered (0.05 M, pH 7.0) Saline (PBS), to the laboratory, within 4 hours. Each sample was dispersed by vorted-mixing and serially diluted. For Aa detection, aliquots from appropriate dilutions were inoculated onto the selective agar media AASM [23] and TSBV [4] modified by omitting the serum (TBV). The diluted sample was also inoculated on the non-selective medium Brucella agar [24]. All agar plates were incubated in air with 5% CO2, at 37°C, for 3 days, and examined under a stereoscopic microscope. On the selective media, all catalase positive (tested with a drop of 3% H2O2) colonies with characteristic morphology, i.e. small (˜1mm in diameter) translucent with rough surface and an internal star-shaped formation [4], were counted. The detection limit of the cultural method was at 50 Colony-Forming Units (CFU) per sample. On Brucella agar, all colonies were counted to determine the total number of facultatively anaerobic bacteria of each sample.
From the agar plates, up to five colonies tentatively identified as Aa, were isolated and cultured on Brucella agar to ensure purity before being subjected to further genetic characterization.
Genetic analysis of isolates
DNA was extracted from pure cultures by boiling a cell suspension of each isolate in distilled water for 10 min. The boiled suspension was placed in an ice bath for 5 min centrifuged to remove cell debris and the supernatant was used as template for PCR analysis. Species identification was accomplished with species-specific primers for 16S rDNA [25] and for the leukotoxin gene [26].
The assay conditions were as described elsewhere [25,26]. Briefly, the PCR mixture contained 2 μl of each primer, 2 μl of dNTP mixture, 10X Ex taq Buffer, 0.1 μl of Takara Ex taq (Takara Biomed., Shiga, Japan) and 5 μl of template DNA in a 20 μl reaction volume. PCR reactions were carried out in a DNA thermal cycler (Applied Biosystems 2720 Thermal Cycler; Applied Biosystems., CA, USA).
The PCR running condition included an initial denaturation at 94°C for 3 min followed by 36 cyclesconsisting of 94°C for 45 s, 55°C for 30 s, 72°C for 45 s and a final extension at 72°C for 10 min. PCR products were analysed by 2.0% agarose gel electrophoresis. A 100-bp DNA ladder (Takara Biomed., Shiga, Japan) was used as a molecular size marker.
To detect the characteristic for the JP2 (highly leukotoxic) clone deletion of 530 bp in the leukotoxin promoter, PCR was run using leukotoxin promoter gene primers as previously described [26]. Under these conditions DNA templates from strains belonging to the JP2 clone give a 504-bp amplicon [27], while DNΑ from all other strains produces a 1034-bp amplicon (Figure 1). The PCR mixture was as described above and the running condition included an initial denaturation at 94°C for 5 min followed by 35 cycles consisting of 94°C for 1 min, 32°C for 2 min, 72°C for 2 min and a final extension at 72°C for 5 min.
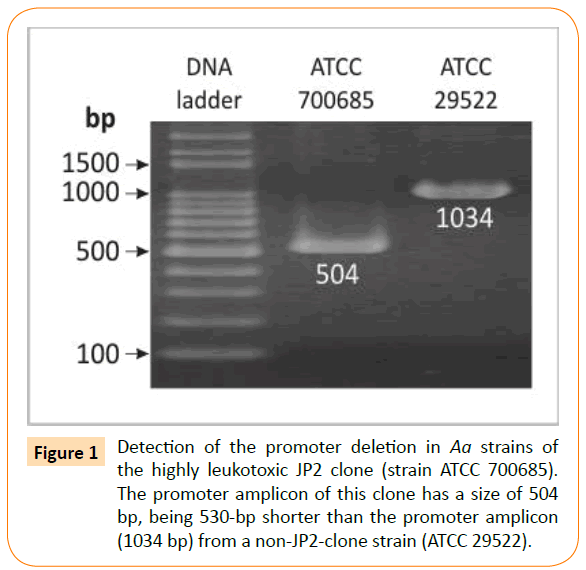
Figure 1: Detection of the promoter deletion in Aa strains of the highly leukotoxic JP2 clone (strain ATCC 700685). The promoter amplicon of this clone has a size of 504 bp, being 530-bp shorter than the promoter amplicon (1034 bp) from a non-JP2-clone strain (ATCC 29522).
Serotyping of the isolates was performed by a serotype-specific PCR assay using O-polysaccharide antigen cluster primers [28]. DNA from the Aa strains ATCC 29523, ATCC 29522, ATCC 33384, IDH 781, OMZ 534, and MUM-A.a 5005 was used as the standards for serotypes a, b, c, d, e, and f, respectively [28]. Electrophoretic separation and detection of the PCR products were performed as described earlier [28]. A typical pattern of serotype specific amplicons is shown in (Figure 2). The PCR mixture contained 0.5 μl of each primer, 2 μl of dNTP Mixture, 10X Ex taq Buffer, 0.1 μl of Takara Ex taq (Takara Biomed., Shiga, Japan) and 5 μl of template DNA in a 20 μl reaction volume. The running condition included 30 cycles (94°C for 30 s and 55°C for 30 s).
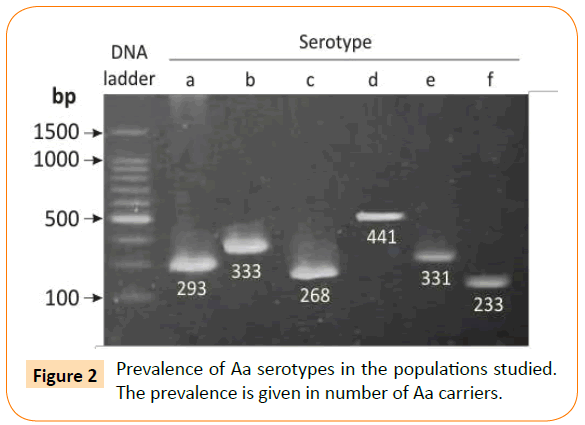
Figure 2: Prevalence of Aa serotypes in the populations studied. The prevalence is given in number of Aa carriers.
To determine a possible strain transmission between children and their mothers the genotype of the isolates was examined by Arbitrarily Primed PCR (AP-PCR) using the random sequence primer OPA 3 and OPA 13 [29]. The PCR mixture and running conditions were the same as for species identification described above.
Statistical Analysis
Statistical results are presented as absolute and relative frequencies (%), minimum, median and maximum values. Mann Whitney test was used to compare distributions relative to their central tendency (median values). Chi squared test was used for comparing percentages and frequency distributions. In all non-parametric statistical tests the observed significance level (P value) was computed either by the Monte-Carlo simulation method (utilizing 10,000 re-sampling circles) or by the exact method. These methods allow drawing safe inductive conclusions even in cases where the required methodological assumptions of the corresponding tests (e.g. large samples, independent measurements, symmetrical distributions, absence of heavy outliers) are not met [30]. In all hypotheses testing procedures, the significance level was predetermined at a=0.05. Statistical analyses was done using the software SPSS v.15.0 (SPSS Inc, Chicago, Illinois, USA) enhanced with the module Exact Tests (for Monte-Carlo and Exact method computations)
Results
The total number of cultivable facultatively anaerobic microbes in the plaque samples from children ranged from 7 × 103 to 5 × 107 CFU/ml sample with a median value of 1 × 106 CFU/ml. The corresponding numbers for the samples from mothers were in the range of 2 × 105 to 9 × 107 CFU/ml and the median value was 1 × 106 CFU/ml.
About 13% of the children, i.e. 14 out of 108, and 23% of the mothers, i.e. 19 out of 83, were found to carry Aa in their supragingival plaque. Aa comprised a minor part of the microflora (Figure 3). The median proportion for the children was 0.087% (min-max: 0.002-4.8%) and for the mothers 0.005% (min-max: 0.001-6.667%). Aa detection was accomplished with the selective culture media TBV and AASM (Table 1). In 23 out of 33 Aa positive samples, Aa was found on both media. In 4 samples, Aa was found only on TSBV and in 6 samples only on AASM. Yet, in 13 samples, a 10-fold higher number of Aa colonies grew on AASM compared to TBV. The opposite finding was recorded in 4 of the Aa-containing samples (Table 1). As expected, a greater number of non-Aa colonies often grew on TBV than on AASM.
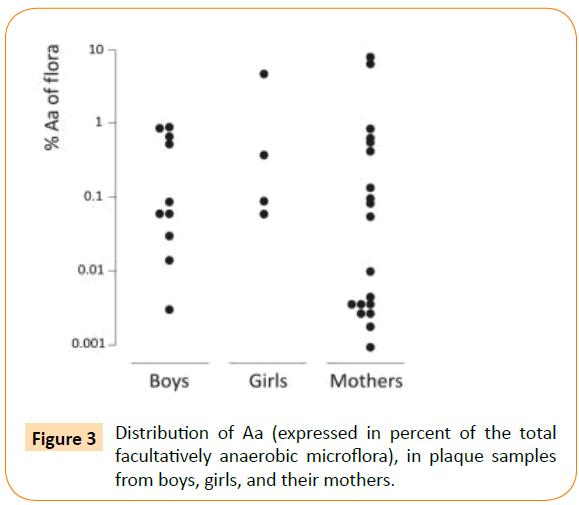
Figure 3: Distribution of Aa (expressed in percent of the total facultatively anaerobic microflora), in plaque samples from boys, girls, and their mothers.
| TBV |
AASM |
| + |
- |
+ >10 |
| + |
23 |
4 |
13 |
| - |
6 |
158 |
|
| + >10 |
4 |
|
|
Table 1: Aa detection by the two selective culture media TBV and AASM. The numbers of dental plaque samples with (+) or without (-) Aa colonies on each medium are given. >10: A 10-fold difference in colony numbers between the media.
Aa was found in 4 out of 108 mother/child pairs. Three pairs had the same serotype, one pair had serotype a and the other two pairs had serotype c. The genetic similarity of the isolates from each mother and her child was established by AP-PCR (Figure 4). From the fourth couple, the strains isolated from the mother and the child were serotype a and c, respectively. Sixteen mothers had more than one child that participated in the study. Eleven mothers had 2 children, 4 had 3, and 1 had 4 children. Among the four siblings, only one, the youngest child, was Aa carrier. In one of the families with three children, the mother and one child harbored Aa in their supragingival plaque.

Figure 4: Genetic similarity (detected by AP-PCR using the primers OPA 13 and OPA 3) of Aa isolates from two child-mother pairs. Strains 8 and 270 were isolated from the children and strains 11 and 273 from their mothers. Strains 81 and 268 (negative control) were isolated from nonrelated children.
Aa strains isolated from the children belonged to serotypes a, b, and c (Table 2). A similar serotype distribution was found in the samples from the mothers (Chi squared P=0.965). In addition, two strains one of each serotype e and f were detected in mothers. For both children and mothers, serotype b was the most prevalent, although serotype c was almost as common as serotype b in mothers (Table 2). None of the strains in the examined population belonged to JP2 clone. No statistically significant difference (Chi squared P=0.554) in serotype distribution was found between boys and girls. Neither was the Aa prevalence related to the age of children (Mann-Whitney P=0.148).
| Serotype |
Children |
Mothers |
| a |
2 |
4 |
| b |
7 |
7 |
| c |
5 |
6 |
| d |
0 |
0 |
| e |
0 |
1 |
| f |
0 |
1 |
| g |
0 |
0 |
| Total |
14 |
19 |
Table 2 Prevalence of Aa serotypes in the populations studied. The prevalence is given in number of Aa carriers.
Discussion
Based on the present results, a considerable number of periodontally healthy Greek children appear to harbor Aa in their supragingival plaque flora. The proportion of Aa carriers, 13%, is far different from those previously reported for Greek populations [21,22], but is close to the ones found in most other Caucasian groups [31,32].
Several methodological factors, such as the sample material and transport, the culture conditions and the detection-identification techniques, may have affected the results of various studies. Based on earlier observations that indicated tooth surfaces as the main colonization site of Aa in the oral cavity [33,34], dental plaque was chosen as the sample material. A culture-based detection technique was also preferred to enable isolation of Aa strains for further characterization. Culture techniques are often less sensitive than molecular methods in detecting small quantities of bacteria. The culture procedure with the selective media TBV and AASM presently used, has a detection limit of 50 CFU/ml per sample, which was considered satisfactory for the aim of the study.
Although more sensitive in detecting small bacteria numbers, molecular methods based on reactions of whole genomic DNA, as in the case of checkerboard technique used elsewhere [22], may sometimes result in false positive samples due to probe crossreactivity with DNA from other bacteria especially if the probes are not optimized for a bacteria density in the range 104-107 in the sample [35]. Furthermore, the technique was developed for subgingival plaque samples and it has to be optimized for other biological samples to give reliable results.
The culture medium AASM contains higher antibiotic concentrations and is more selective than TBV [23]. TSBV and TBV have extensively been used for the detection of Aa, mainly in samples from periodontal pockets and recovery of Aa is conveniently accomplished when incubated aerobically and in the presence of 5% CO2 [4,36]. However, when samples with a different flora composition, e.g. supragingival plaque, are cultured, the higher selectivity of AASM might be preferable. The greater recovery of Aa by AASM than TBV in 39% of the samples, mainly due to a lessen growth of non-Aa colonies, compared to TSBV, supports the above aspect.
Intra-familial spread of Aa, often from parents to their children, has previously been suggested [9,10]. On the other hand, Aa has been isolated from children whose parents lacked the bacterium in their oral flora [10]. Genotypic identity of isolates between mothers and their children has been observed in about 30% of the cases [9]. Although at lower frequency, genotypic similarity of Aa isolates between mothers and children was also observed in the present study. However, it seems that acquisition of Aa by children mostly happens through contamination from other sources since most of the children harbored Aa genotypes that were different were different from those in their mothers. The possibility for intra-familiar contamination cannot be excluded to occur beside the one from other sources. However, contamination is not the only requirement to be fulfilled for a bacterium to colonize a specific environment. Several ecological factors may affect bacterial colonization in a particular habitat. As presently revealed, Aa is not always found in all siblings, while different genotypes are detected among children and their mothers, this being in line with previous results [9,10]. Probably, the conditions prevailing in the oral environment of the host have to match the colonization requirements of each Aa genotype and various genotypes may be adapted to certain hosts. Host defense, bacterial antagonism, and possibly lack of pathogenicity of infecting organisms may influence transmission. Periodontal pathogens are suggested to be communicable; however, they are not readily transmissible [12]. There are conflicting opinions as to whether certain putative periodontal pathogens such as Aa are exogenous or endogenous that have particular characteristics. If periodontal infections result from an overgrowth of endogenous bacteria, then transmission is not considered a critical issue [12].
The increased prevalence of serotypes b and c observed in this child population is generally in agreement with the results from other studies with Caucasian groups [37-40]. In mothers, serotype b was the most prevalent and serotype c almost equally frequent, this being in line with the results from an earlier study showing serotype c to be the most frequently encountered Aa serotype in periodontal pockets of adults from the same geographic region [37].
Despite the high prevalence of serotype b strains in this population, none of the isolates belonged to the strongly leukotoxic clone JP2. This finding is in accordance with previous results that support the aspect of a racial tropism for this specific clone and its absence from the oral cavity of Caucasians [17,41,42]. However this does not exclude the possibility of occurrence of other highly leucotoxic strains of Aa as previously reported [43].
In young people, a relation was suggested between the subject's age and the colonization of oral cavity with Aa [3]. The present study as much as others [44], failed to confirm this relation and also showed that children may carry periodontal pathogens from an early age. However, the non-longitudinal type of this study and the relatively small number of Aa-carrying children may have been responsible for not detecting a possible relation between the child's age and the Aa occurrence in the mouth.
To conclude, the prevalence of Aa and its serotype distribution pattern, in Greek children, are similar with those found in other Caucasians. The occurrence of Aa in dental plaque of children and their mothers infrequently coincides.
Acknowledgements
The authors would like to acknowledge the pediatric dentists of the social Insurance Institution of Thessaloniki Dr. Vlachou Ioanna and Dr. Karafergia Euterpi and the members of the department of Pediatric Dentistry of Aristotle University of Thessaloniki for their valuable assistance.
7475
References
- Johansson A, Kalfas S (2012) Virulence Mechanisms of Leukotoxin from Aggregatibacteractinomycetemcomitans. In: Mandeep SV, Oral Health Care - Prosthodontics, Periodontology, Biology, Research and Systemic Conditions. InTech 165-192.
- DiRienzo JM, McKay TL (1994) Identification and characterization of genetic cluster groups of Actinobacillusactinomycetemcomitans isolated from the human oral cavity. J ClinMicrobiol 32: 75-81.
- Yuan K, Hsu PC, Tseng CC, Kiang D, Wang JR (2001) Detection rate of Actinobacillusactinomycetemcomitans on the permanent 1st molars of primary school children in Taiwan by polymerase chain reaction. J ClinPeriodontol 28: 348-352.
- Slots J (1982) Selective medium for isolation of Actinobacillusactinomycetemcomitans. J ClinMicrobiol 15: 606-609.
- Rotimi VO, Salako NO, Divia M, Asfour L, Kononen E (2010) Prevalence of periodontal bacteria in saliva of Kuwaiti children at different age groups. J Infect Public Health 3: 76-82.
- Ebersole JL, Holt SC, Delaney JE (2014) Acquisition of oral microbes and associated systemic responses of newborn nonhuman primates. Clin Vaccine Immunol21:21-28.
- Petit MD, Van Steenbergen TJ, De Graaff J, Van der Velden U (1993) Transmission of Actinobacillusactinomycetemcomitans in families of adult periodontitis patients. J Periodontal Res 28: 335-345.
- Dogan B, Kipalev AS,Okte E, Sultan N, Asikainen SE (2008) Consistent intrafamilial transmission of Actinobacillusactinomycetemcomitans despite clonal diversity. J Periodontol 79:307-315.
- Pahkla ER, Jogi E, Nurk A, Pisarev H, Koppel T, et al. (2010)Periodontal disease in mothers indicates risk in their children. Int J Paediatr Dent 20: 24-30.
- Tamura K, Nakano K, Hayashibara T, Nomura R, Fujita K, et al. (2006) Distribution of 10 periodontal bacteria in saliva samples from Japanese children and their mothers. Arch Oral Biol 51: 371-377.
- Rego RO, Spolidorio DM, Salvador SL, Cirelli JA (2007) Transmission of Aggregatibacteractinomycetemcomitans between Brazilian women with severe chronic periodontitis and their children. Braz Dent J 18:220-224.
- Greenstein G, Lamster I (1997) Bacterial transmission in periodontal diseases: a critical review. J Periodontol 68:421-431.
- Yang HW, Huang YF, Chan Y, Chou MY (2005) Relationship of Actinobacillusactinomycetemcomitans serotypes to periodontal condition: prevalence and proportions in subgingival plaque. Eur J Oral Sci 113: 28-33.
- Roman-Torres CV, Aquino DR, Cortelli SC, Franco GC, Dos Santos JG, et al. (2010) Prevalence and distribution of serotype-specific genotypes of Aggregatibacteractinomycetemcomitans in chronic periodontitis Brazilian subjects. Arch Oral Biol 55: 242-248.
- Pinheiro ET, Kawamoto D, Ota-Tsuzuki C, Almeida LR, Nunes AC, et al. (2011) Analysis of genotypic variation in genes associated with virulence in Aggregatibacteractinomycetemcomitans clinical isolates. J Periodontal Res 46: 310-317.
- Takada K, Saito M, Tsuzukibashi O, Kawashima Y, Ishida S, et al. (2010) Characterization of a new serotype g isolate of Aggregatibacteractinomycetemcomitans. Mol Oral Microbiol 25: 200-206.
- Haubek D, Dirienzo JM, Tinoco EM, Westergaard J, López NJ, et al. (1997) Racial tropism of a highly toxic clone of Actinobacillusactinomycetemcomitans associated with juvenile periodontitis. J ClinMicrobiol 35: 3037-3042.
- Asikainen S, Chen C (1999) Oral ecology and person-to-person transmission of Actinobacillusactinomycetemcomitans and Porphyromonasgingivalis. Periodontol 2000 20: 65-81.
- Belibasakis GN, Mattsson A, Wang Y, Chen C, Johansson A (2004)Cell cycle arrest of human gingival fibroblasts and periodontal ligament cells by Actinobacillusactinomycetemcomitans: involvement of the cytolethal distending toxin. Apmis 112: 674-685.
- Lally ET, Hill RB, Kieba IR, Korostoff J (1999)The interaction between RTX toxins and target cells. Trends Microbiol7: 356-361.
- Kamma JJ, Diamanti-Kipioti A, Nakou M, Mitsis FJ (2000) Profile of subgingivalmicrobiota in children with mixed dentition. Oral MicrobiolImmunol 15: 103-111.
- Papaioannou W, Gizani S, Haffajee AD, Quirynen M, Mamai-Homata E (2009) The microbiota on different oral surfaces in healthy children. Oral MicrobiolImmunol 24: 183-189.
- Tsuzukibashi O, Takada K, Saito M, Kimura C, Yoshikawa T, et al.( 2008) A novel selective medium for isolation of Aggregatibacter (Actinobacillus) actinomycetemcomitans. J Periodontal Res 43: 544-548.
- Kaklamanos EG, Charalampidou M, Menexes G, Topitsoglou V, Kalfas S (2005) Transient oral microflora in Greeks attending day centres for the elderly and residents in homes for the elderly. Gerodontology 22: 158-167.
- Ashimoto A, Chen C, Bakker I, Slots J (1996) Polymerase chain reaction detection of 8 putative periodontal pathogens in subgingival plaque of gingivitis and advanced periodontitis lesions. Oral MicrobiolImmunol 11: 266-273.
- Tønjum T, Haas R (1993) Identification of Actinobacillusactinomycetemcomitans by leukotoxin gene-specific hybridization and polymerase chain reaction assays. J ClinMicrobiol 31: 1856-1859.
- Haubek D, Poulsen K, Westergaard J, Dahlèn G, Kilian M (1996) Highly toxic clone of Actinobacillusactinomycetemcomitans in geographically widespread cases of juvenile periodontitis in adolescents of African origin. J ClinMicrobiol 34: 1576-1578.
- Kaplan JB, Perry MB, MacLean LL, Furgang D, Wilson ME, et al.(2001) Structural and genetic analyses of O polysaccharide from Actinobacillusactinomycetemcomitans serotype f. Infect Immun 69: 5375-5384.
- Paju S, Saarela M, Alaluusua S, Fives-Taylor P, Asikainen S (1998) Characterization of serologically non-typeableActinobacillusactinomycetemcomitans isolates. J ClinMicrobiol 36: 2019-2022.
- https://priede.bf.lu.lv/grozs/Datorlietas/SPSS/SPSS%20Exact%20Tests%207.0.pdf
- Kulekci G, Leblebicioglu B, Keskin F, Ciftci S, Badur S (2008)Salivary detection of periodontopathic bacteria in periodontally healthy children. Anaerobe 14:49-54.
- Fine H, Kaplan JB, Kachlany SC, Schreiner HC (2006)How we got attached to Actinobacillusactinomycetemcomitans: a model for infectious diseases. Periodontology 2000 42: 114–157.
- Mager DL, Ximenez-Fyvie LA, Haffajee AD, Socransky SS (2003) Distribution of selected bacterial species on intraoral surfaces. J ClinPeriodontol 30: 644-654.
- Kilian M, Schiott CR (1975)Haemophili and related bacteria in the human oral cavity. Arch Oral Biol 20: 791-796.
- NacimentoC, Issa MJP, Watanabe E, Ito YI (2006) DNA checkerboard method for bacterial pathogen identification in oral diseases. Int J Morphol24: 619-624.
- Holm A,Rabe P, Kalfas S, Edwardsson S (1987) Improved selective culture media for Actinobacillusactinomycetemcomitans and Haemophilusaphrophilus. J ClinMicrobiol 25:1985-1988.
- Sakellari D, Katsikari A, Slini T, Ioannidis I, Konstantinidis A, et al. (2011) Prevalence and distribution of Aggregatibacteractinomycetemcomitans serotypes and the JP2 clone in a Greek population. J ClinPeriodontol 38: 108-114.
- Hölttä P, Alaluusua S, Saarela M, Asikainen S (1994) Isolation frequency and serotype distribution of mutans streptococci and Actinobacillusactinomycetemcomitans, and clinical periodontal status in Finnish and Vietnamese children. Scand J Dent Res102: 113-119.
- Haubek D, Poulsen K, Asikainen S, Kilian M (1995) Evidence for absence in northern Europe of especially virulent clonal types of Actinobacillusactinomycetemcomitans. J ClinMicrobiol 33: 395-401.
- Teixeira RE, Mendes EN, Roque de Carvalho MA, Nicoli JR, FariasLde M, et al. (2006)Actinobacillusactinomycetemcomitans serotype-specific genotypes and periodontal status in Brazilian subjects. Can J Microbiol 52: 182-188.
- Macheleidt A, Müller HP, Eger T, Putzker M, Fuhrmann A, et al. (1999) Absence of an especially toxic clone among isolates of Actinobacillusactinomycetemcomitans recovered from army recruits. Clin Oral Investig 3: 161-167.
- Sakai VT, Campos MR, Machado MA, Lauris JR, Greene AS, et al. (2007) Prevalence of four putative periodontopathic bacteria in saliva of a group of Brazilian children with mixed dentition: 1-year longitudinal study. Int J Paediatr Dent 17: 192-199.
- Höglund-Åberg C, Haubek D, Kwamin F, Johansson A, Claesson R (2014)Leukotoxic activity of Aggregatibacteractinomycetemcomitans and periodontal attachment loss. PLoS ONE 9: e104095.
- Lamell CW, Griffen AL, McClellan DL, Leys EJ (2000) Acquisition and colonization stability of Actinobacillusactinomycetemcomitans and Porphyromonasgingivalis in children. J ClinMicrobiol 38: 1196-1199.









