Keywords
Gastric biopsies; Helicobacter pylori; Histology; Urease; Reliability Immunohistochemistry
Introduction
Helicobacter pylori is a Gram-negative microaerophilic spiral bacterium discovered in 1983 [1]. It infects more than half of the world’s human population with prevalence ranging from 25% in developed countries to more than 90% in developing countries [2]. The increased risk of infection is especially high among those living in the developing world [3]. The principal reasons for these variations may involve socio-economic differences between populations. A lack of proper sanitation, safe drinking water, and basic hygiene as well as poor diets and overcrowding all play a role in determining the overall prevalence of infection [4]. Infection with the bacterium causes chronic gastritis, peptic ulceration, gastric cancers and gastric Mucosa Associated Lymphoid Tissue (MALT) Lymphoma [3]. Helicobacter pylori has been rated as a “class one” carcinogen to the gastrointestinal tract by the World Health Organization [5]. It is in the same category as cigarette smoke is to lung cancer.
It has been suggested that up to 95% of duodenal and 70% of gastric ulcers are attributed to Helicobacter pylori infection [6]. Most cases occur among middle aged subjects and the highly productive age groups in societies 6. Infections can commence early in life and are lifelong if remedial actions are not taken [7].
Helicobacter pylori almost exclusively colonizes the mucous layer of the human stomach. Once established, the pathogen may reside in the host for decades. However, in 10% of cases, the organism is associated with diverse clinical outcomes, which include non-ulcer dyspepsia (NUD), peptic ulcer disease (PUD), and gastric cancer [8]. Several host and bacterial virulence factors have been related to this clinical diversity.
Helicobacter pylori is believed to be transmitted primarily by faecal oral or oral-oral routes, with water and food as possible vehicles of infection. However, exact modes of transmission are not easily determined because Helicobacter pylori is difficult to culture from environmental samples. There is some evidence for iatrogenic transmission through inadequately sterilized endoscopes. Helicobacter pylori has been detected in vomitus, indicating the potential for gastro-oral transmission [9].
The prevalence is generally high in developing countries like Nigeria. Prevalence of 82% has been reported in children 5-9 years, 95% in adults of middle age and 70–90% in older adults 4. There is an estimate of more than 80% of Africans infected with Helicobacter pylori [10]. A study on sero-prevalence of Helicobacter pylori infected patients with peptic ulcer in Kaduna State of Nigeria revealed that out of the 225 patients tested, 181 were negative [11]. A similar study was carried out in Enugu state, Nigeria where out of 103 patients, 63 (62%) were positive [12]. Ranked as a class one carcinogen, Helicobacter pylori continues to present itself as a serious health concern [5]. However, no literature has been found on its prevalence in Benue State, Norther Central Nigeria. (80.4%) were positive for Helicobacter pylori and 44 (19.6%)
The burden of Helicobacter pylori infection is so much that infected individuals live the rest of their lives taking drugs, avoiding certain foods and drinks because they believe it has no cure [13]. Although extensive research has been carried out on Helicobacter pylori, the research so far in Nigeria has focused on its prevalence in certain parts/states and very little literature was found on testing the reliability of the different methods of detecting the bacterium.
Helicobacter pylori is very common in Nigeria as in other developing countries [14]. Disease Outcome may be a result of several factors including host factors, environmental factors and differences in the prevalence or expression of bacterial elements [15]. Since the discovery of Helicobacter pylori as an important etiological agent in gastritis and peptic ulcer disease, investigation for this bacterium during endoscopy has become a standard clinical practice to establish active Helicobacter pylori infection [16]. In Nigeria, there is considerable confusion over the management of Helicobacter pylori infection. There are no national consensus guidelines and very few documented pathological role of the bacterium [17]. There are various techniques of detecting Helicobacter pylori from specimens. These tests may be invasive or non-invasive [18]. Endoscopy and gastric mucosal biopsy, microscopic examination of histological sections and rapid urease test are forms of invasive test that could be used. Non-invasive tests such as Urea Breath Test (UBT) make use of the ability of the organism to produce urease; enzyme linked Immunosorbent assay (ELISA), Helicobacter pylori stool antigen test and latex agglutination tests are important non- invasive serological approaches employed to detecting presence of antibody or antigen from a specimen [19]. As such this study is aimed at assessing the reliability of PCR, microbiology and histologic methods in detecting Helicobacter pylori in gastric biopsies of patients referred for endoscopy at Benue State University Teaching Hospital Makurdi.
Sample collection
Gastric biopsy samples were taken from the antrum of patients referred for endoscopy at Benue State University Teaching Hospital Makurdi who had not been on any antibiotic or eradication therapy for the past four months. Endoscopies were performed by Gastroenterologists using standard endoscopy procedures. The tissue samples collected were put in two different bottles, a sterile McCartney bottle containing Brain Heart Infusion broth for microbiology tests and a plastic universal bottle containing 10% formal saline for histopathological examinations.
Tissue processing
The biopsy tissues in 10% formal saline were processed for histopathology using an automated tissue processor (ATP). Formalin Fixed Paraffin Embedded (FFPE) tissue blocks were sectioned using a microtome, cut into thin sections of 4 micrometre thickness, dewaxed and used for histopathological staining.
Haematoxylene and eosin staining (H&E)
Dewaxed and cleared slide sections were stained in Haematoxylene for 5 minutes and rinsed in 1% acid alcohol for a few seconds. The sections were blued in tap water and counterstained in Eosin solution for 5 minutes and washed in tap water. The sections were dehydrated in ethanol (70%, 95%, and 100%) for 3 minutes each and mounted in Dextrene plasticizer xylene (DPX) to increase refractive index.
Immunohistochemical staining (IHC)
Mouse and Rabbit Specific HRP/DAB detection kit from ABcam (Cambridge, UK; Cat No. ab64264) was used for immunostaining.
Slides were labelled clearly and arranged in a staining rack including two slides for positive and negative controls. The negative control was a known gastric biopsy without Helicobacter pylori while the positive control was a section known to have Helicobacter pylori. The immedge hydrophobic pen was used to carefully mark the area of slide staining. Tissue sections were blocked in hydrogen peroxide and incubated at room temperature in a humid chamber for 10 minutes followed by washing two times in PBS buffer with 1% Tween 20 (Wash Buffer) for three minutes each. About 80 μl of protein block was applied to cover sections and incubated at room temperature in a humid chamber for 5 minutes and washed once in Wash Buffer for three minutes. About 80 μl of primary antibody (specific to the cellular antigen), unconjugated Helicobacter pylori antibody (from Fitzgerald Industries, USA) was applied onto sections and incubated at room temperature in the humid chamber for 40 minutes then washed four times in the Wash buffer for three minutes each. Anti-mouse and Rabbit (Biotinylated goat) 80 μ1 was applied and incubated at room temperature for 10 minutes in a humid chamber and washed four times in buffer for three minutes each followed by addition of 80 μl of streptavidin peroxidase and incubation at room temperatures for ten minutes in the humid chamber. The slides were rinsed four times in Wash Buffer. DAB solution 100 μl was added to the slides and incubated at room temperature for 10 minutes then rinsed four times in the buffer. Filtered Haematoxylin 100 μl (counterstain) was added onto slides and incubated at room temperature for one minute.
Slides were rinsed 7 times in tap water, dehydrated, cleaned in xylene and mounted, each slide with DPX mountant avoiding air bubbles.
Giemsa stain for tissue sections
Tissue sections were stained in Haematoxylene stain for 5 minutes. They were rinsed in water and placed in 1% acid alcohol for a few seconds. The sections were blued in tap water.
They were then placed in Giemsa stain for 5 minutes and washed in tap water.
The sections were dehydrated in ethanol (70%, 95%, and 100%) for 10 minutes each and mounted in DPX to increase refractive index (clear). Giemsa stain helps to demonstrate the presence of the organism.
Helicobacter pylori antigen enzyme linked immunosorbent assay (ELISA)
Diagnostic Automation ELISA Helicobacter pylori antigen kit Lot No. 1LD5-213 was used to detect Helicobacter pylori antigens in the gastric biopsy specimens.
The biopsy specimen in brain heart infusion broth with 1.5% glycerol was briefly mixed by vortexing to homogenize the specimen. About 100 μl of sample was dispensed into wells already coated with Helicobacter pylori antibody. It was swirled gently to mix and expel bubbles and incubated at room temperature for 30 minutes. The liquid was removed from wells and washed three times with washing buffer (phosphate buffered saline with Tween 20) using a multichannel pipette.
100 μl of enzyme conjugate containing specific antibody for Helicobacter pylori and an enzyme (Horse Radish Peroxidase HRP) was added and incubated for 30 minutes at room temperature. The wells were washed three times in washing buffer. 100 μl of Tetra methyl benzathene (TMB) chromogenic substrate was added and incubated at room temperature for 30 minutes.
100 μl of stop solution (Hydrochlonic acid) was added to stop the reaction. Intense yellow indicates a positive result. The solutions were read at 450 nm using Mindray 96 well Microplate Reader.
Extraction of genomic DNA (gDNA)
Genomic DNA was extracted from the tissue samples using ReliaPrep genomic DNA miniprep kit (Promega, Southampton UK). The ReliaPrep uses spin columns that contain silica membrane for DNA purification. Briefly, about 200 μl of the macerated tissue materials in broth were dispensed into 2 ml Eppendorf tube containing 25 μl of proteinase K. The sample was mixed by gentle vortex and incubated for 10 minutes at room temperature. Then 200 μl of Cell Lysis Buffer was added and the sample vortexed for 10 seconds before incubation in a water bath set at 56°C for 10 minutes. Thereafter, 250 μl of Binding Buffer was added to the sample and mixed by repeated pipetting. The mixture was transferred to the Spin Column and centrifuged at 14000 rpm for one minute. The flow through in the collection tube was discarded. The column was washed by addition of 500 μl of Column Wash Buffer and centrifuged for 3 minutes at 14000 rpm. The washing was repeated twice. Columns were then placed into new Collection tubes and centrifuged at 14000 rpm for 1 minute to remove residual Wash Buffer. Then 100 μl of Nuclease-free water was added into the columns, which were placed into 1.5 ml tubes, incubated for one minute at room temperature and centrifuged at 13000 rpm for one minute. DNA quality was checked by reading at 260/280 nm using Eppendorf Biophotometer Plus (Eppendorf, Germany). The DNA elute was labeled and stored in the fridge until required for testing.
Specific primers for detection of Helicobacter pylori
The primer sequences used were: 16S rRNA- F (forward): GGAGGATGAAGGTTTTAGGATTG,
16S rRNA-R = TCGTTTAGGGCGTGGACT [20,21]. Primers were synthesized by Eurofins, Germany.
Detection of H pylori 16S rRNA gene
Stock primers were dissolved in 1x TE solution at pH 8.0 and stepped down to 10 μM working primer. PCR was set up in a PCR plate using 12.5 μl of the PCR master mix, 7.5 μl of the primer mix (at final concentration of 500 Nm) and 5.0 μl of genomic DNA. Thermal profile was 95°C for two minutes for initial denaturation, 40 cycles of 94°C for 18 seconds, 60°C for 30 seconds, 72°C for 30 seconds and final extension of 72°C for 5 minutes using Eppendorf Nexus Gradient Master Cycler. It amplified 294 bp product sizes.
PCR products were electrophoresed in 2.0% agarose gel at 100 v for 30 minutes Images were captured using Geno-Mini Electrophoresis Gel system. (VWR, UK).
Detection of Helicobacter pylori using SYBR Green Real Time PCR
The 2x SYBR master mix (Promega, Southampton, UK) was used for this reaction. This is a Real-Time PCR designed to detect both culturable and non-culturable forms of H pylori using Melting curve analysis. It is based on 16S rRNA gene and the primer system amplifies 294 bp product size using a final concentration of 0.5 μM in a 25 μL reaction volume. The thermal profile comprised of initial denaturation at 95°C for 3 mins followed by 35 cycles of 94°C for 30 secs, 70°C for 60 secs, 72°C for 60 secs, and one cycle for melt curve of 94°C for 30 sec and final extension of 72°C for 30 sec. The Real-Time PCR was performed on ABI One Step Real Time PCR system. The melting point for positive amplified product for Helicobacter pylori was 72°C.
The primer sequences used were: HP294F AAGCTT TTAGGGGTGTTAGGGGTTT-HP294R AAGCTTACTTTCTAACACTAACGC- Chamanrokh, et al. (2015); Modified
Urease test for detection of Helicobacter pylori
Urea Agar Base (Oxoid) was used.
2.4% of urea agar base was suspended in of distilled water and brought to boil to dissolve completely.
This was sterilized by autoclaving at 115°C for 20 minutes and cooled to 50°C One ampule of 40% sterile urea solution was added aseptically and mixed well 10 ml amounts were distributed into sterile tubes and allowed to set in the slope position and the gastric biopsy samples in brain heart infusion broth were stab inoculated in the urease agar, covered with cotton wool and incubated for 16 hours.
Positive samples changed the colour of the agar from yellow to purple-pink.
Culture of gastric samples
The gastric biopsy specimens in Brain Heart Infusion broth containing 1.5% glycerol were homogenized and cultured on commercially prepared Helicobacter pylori Selective Agar PB0398A from Oxoid (Basingstoke, UK) and incubated at 37°C in a microaerophilic condition using GENbox microaerophilic packs (Biomerieux, UK) for 5 days. Small, circular, smooth colonies observed were Gram stained to check for Gram reaction. Colonies were also examined for urease, catalase and oxidase reactions.
Gram stain: A loopful of colonies was placed on a clean slide, a drop of sterile water added and emulsified. Another slide placed at an angle on it and a smear/thin film made by dragging the slide over it. The smear was air dried and heat fixed by passing over a flame. It was first immersed in crystal violet for one and a half minutes. Water was used to wash the slide and then immersed in Gram ?s iodine for one minute. The smear was then washed with water and decolourised with acetone till the violet colour was cleared. Dried slides were examined at X100 objective using oil immersion.
Gram positive organisms take up the primary stain colour while Gram negative organisms take up the counterstain colour.
Urease test: As earlier described above.
Catalase test: A drop of three percent hydrogen peroxide was placed on the colonies and prompt effervescence indicated catalase production because catalase degrades hydrogen peroxide and releases oxygen which is detected as effervescence.
Oxidase test: A drop of freshly prepared one percent solution of oxidase reagent was put on a piece of filter paper. The test colony was picked with a sterile loop and rubbed on the filter paper on the area impregnated with the oxidase reagent. Oxidase positive organisms will turn the paper deep purple blue in colour in a few seconds. This indicates the ability of organisms to oxidize amines.
Helicobacter pylori is Gram negative, urease, catalase and oxidase positive.
Statistical analysis: Data were analysed using Statistical Package for Social Sciences (SPSS) version 20, IBM Inc. Chi square was carried out for determination of homogeneity of Helicobacter pylori among the subjects, Kappa statistic for interagreement between different methods and PCR method. Alpha level of significance was set at 0.05.
Results
The detection of Helicobacter pylori by different diagnostic methods are shown in Figure 1. Urease had the highest positivity of 91.25% followed by Haematoxylene & Eosin 37%, 16S rRNA PCR 30%, SYBR Green melting curve PCR 27.5%, Immunohistochemical stain 23.75%, Giemsa stain 21.25%, ELISA 5.0% while culture was the least with 1.25%. PCR of 16S rRNA gene was used as the gold standard. Results of statistical comparison of the methods are shown in Table 1. Immunohistochemistry and SYBR Green had almost perfect agreement with PCR (kappa value=0.840; p<0.001, kappavalue= 0.88; p<0.001 respectively), Giemsa had a substantial agreement with PCR (kappa=0.770; p<0.001) ELISA had a fair agreement (kappa-value=0.220; p=0.002) while culture had a weak agreement (kappa-value=0.057; p=0.120).
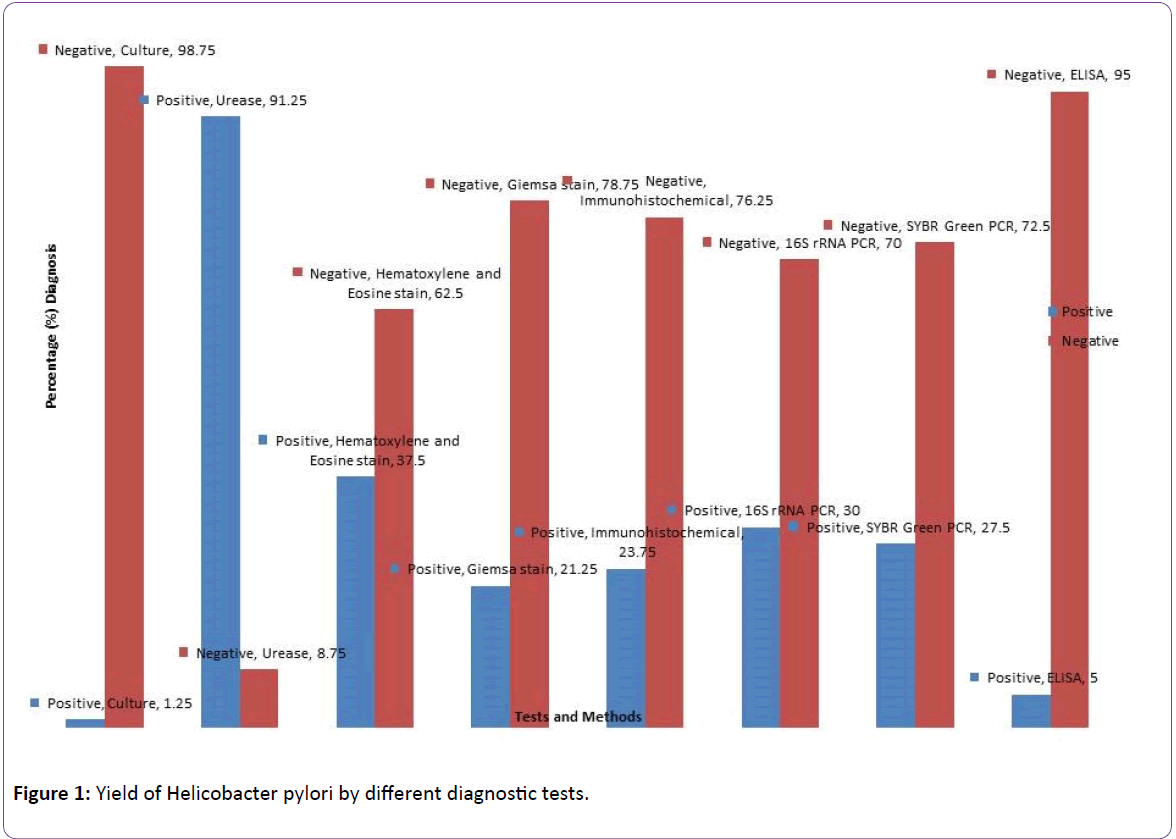
Table 1: Yield of Helicobacter pylori by different diagnostic tests.
| Tests |
Singleplex PCR |
Kappa value |
P-value |
| Positive |
Negative |
| Giemsa stain |
| Positive |
17 |
0 |
|
|
| Negative |
7 |
56 |
0.77 |
<0.001 |
| Immuno-histochemical stain |
| Positive |
19 |
0 |
|
|
| Negative |
5 |
56 |
0.84 |
<0.001 |
| SYBR Green |
| Positive |
21 |
1 |
|
|
| Negative |
3 |
55 |
0.88 |
<0.001 |
| ELISA |
| Positive |
4 |
0 |
|
|
| Negative |
20 |
56 |
0.22 |
0.002 |
| Culture |
| Positive |
1 |
0 |
|
|
| Negative |
23 |
56 |
0.057 |
0.12 |
Table 1: Comparative analysis of diagnostic test for Helicobacter pylori.
<0=less than chance agreement (poor)
0.01-0.20=slight agreement (slight)
0.21-0.40=fair agreement (fair)
0.41-0.60=moderate agreement (moderate)
0.61-0.80=substantial agreement (substantial)
0.81-0.99=almost perfect agreement
Discussion
Apart from PCR, several other methods were used to detect the presence of Helicobacter pylori in biopsy samples to achieve maximum positivity. In the comparative analysis of the methods using kappa statistic, PCR was used as gold standard. PCR has increasingly been described as the gold standard for detecting some microbes [21,22]. Immunohistochemical stain and SYBR Green had an almost perfect agreement with PCR, Giemsa stain had a substantial agreement, ELISA had a fair agreement and culture had a slight agreement. The poor agreement of culture with PCR might be due to the long period of about one year that was used in collection of biopsies, storage and transportation, which could have caused the organism to die despite all the measures taken to preserve it. Anderson also reported low positivity rate of culture in Helicobacter pylori diagnosis and attributed it to loss of viability during transport, low number of organisms, absence of organisms in the gastric biopsies, fastidious growth requirements and presence of non-culturable coccoid forms [23]. Helicobacter pylori may also have a patchy distribution, and the more biopsy specimens analyzed, the higher the chances of detection [24]. The higher sensitivity of Urease test could also be influenced by the presence of other urease positive bacteria in the specimen. Helicobacter heilmanii, which may be present in the stomach, is also a urease-positive bacterium but may be distinguished by histological examination [25].
The availability of different methods for detecting H pylori in gastric biopsies provides choices for eliminating false negative results by using two or more techniques.
Conclusion
All the test methods used in the study had a significant agreement with PCR which was used as the gold standard except ELISA and culture. Combination of two or more techniques for detecting Helicobacter pylori infections in gastric biopsies will help to reduce false negative results and improve clinical management of dyspeptic patient.
Acknowledgements
The research was carried out at the Safety Molecular Pathology Laboratory Enugu, supported in part by a research grant from the Federal University of Agriculture Makurdi, Benue, Nigeria. We also thank the Benue State University Teaching Hospital Makurdi, Nigeria for providing the gastric biopsy specimens and all patients who volunteered to participate.
20849
References
- Warren JR, Marshall B (1983) Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet 1: 1273-1275.
- Bardhan K (1997) Epidemiological Features ofHelicobacter pylori Infection in developing counties. Clinical Infectious Diseases 25:973-879.
- Ahmed KS, Khan AA, Ahmed I, Tiwari SK, Habeeh A, et al. (2007) Impact of Household Hygiene and water source onthe prevalence ofHelicobacter pylori: a South Indian perspective. Singapore medical Journal 48: 543-549.
- Hunt RH, Xiao SD, Mégraud F, Bazzoli F, Van der Merwe S, et al. (2010)Helicobacter pylori in developing countries. World astroenterology Organization Global Guidelines 2-5.
- Aguemon BD, Struelens MJ, Massaoughbodji A, Quendo EM (2005) Prevalence and risk factors forHelicobacter pylori infection in urban and rural Beninese populations. Clinical microbiology and infectious Diseases II 18:611-617.
- Rothenbacher D (2007) IsHelicobacter pylori infection a necessary condition for non-cardia gastric cancer? A view from epidemiology. Arquiros de Medicina21:3.
- Ahmed KS, Khan AA, Ahmed I, Tiwari SK, Habeeb MA, et al. (2006). Prevalencestudy to elucidate the transmission pathways ofHelicobacter pylori at oral andgastroduodenal sites of a South Indian population Singapore medical Journal 47: 291-296.
- Cremonini F, GasbarriniA, Armizzi A, Gasbarrini G (2001)Helicobacter pylori related diseases. European Journal of Clinical Investigation 31: 431-437.
- Tanih NF, Okeleye BI, Naido N, Clarke AM,Mkweshana N, et al. (2010) Marked susceptibility of South AfricanHelicobacter pyloristrainsto ciprofloxacin and amoxicillin: Clinical implication.South Africac Journal of Medicine 100: 49-52.
- CampbellDI, Warren BF, Thomas JE, Figura N, Telford JL, et al. (2010) The African enigma; low prevalence of gastric atrophy, high prevalence of chronicinflammation in west African adults and children. Helicobacter 6: 263-267.
- Nwodo EN, Yakubu SE, Jatau ED, Yaboya A (2009) Seroprevalence ofHelicobacter pylori infection in patients with Gastritisand Peptic Ulcer Disease in Kaduna,Kaduna State, Nigeria. African Journal of Basic and Applied Sciences 1: 123-128.
- Neri GP, Raymond AA, Nora CU, Uzoma CM, Chinyere PN, et al. (2009)Helicobacter pylori prevalence in patients with upper G.I symptoms in Enugu Metropolis.Nigerian Journal of Gastroenterology and Hepatology 1: 37-49.
- Ahuja V, Sharma MP (2002) High recurrence rate ofHelicobacter pylori infection indeveloping countries. Gastroenterology 123: 653-654.
- Ashraf P, Haq MU, Amad R (1999) Assessment ofHelicobacter pylori infectionJournal of Collective Physicians in Surgery Pakistan9:75-77.
- Bani-Hani KE (2002) The status ofHelicobacter pylori. Saudi Medical Journal23: 379-383.
- Ndububa DA, Agbakwura AE, Adebayo RA, Olassode BJ, Olaomi OO et al. (2001) Upper gastrointestinal findings and incidence ofHelicobacter pylori infection among Nigerian patients with dyspepsia. West African Journalof Medicine20: 140-145.
- Adesanya AA, Oyedeje KS, Oluwafowoju IO, Kehinde MO, Coker AO (2003) Diagnosis ofHelicobacter pylori infection in dyspeptic Nigerians: Comparison of UreaseTest, Culture and Serology. Journal of Clinical Science3: 29-34.
- Shepherd JA, Williams LC, Doherty PC, HossackM, Preston T, et al. (2000) Comparison of an enzyme immunoassay for the detection ofHelicobacter pylori antigens in the faeces with the urea breath test. Archives of Diseases inChildren 83:5-13.
- Krogfelt KA, Lehours P, Mégraud F (2005) Diagnosis ofHelicobacter pylori infection. Helicobacter 10: 5-13.
- Chamanrokh P, Mohammad HS, Mahnaz MS, Taher N, Davood, E (2015) Three TestsUsed to Identify Non-culturable Forms ofHelicobacter pylori in Water Ssamples. Journal ofMicrobiology 8: 16811.
- Mackay WG, Williams CL, McMillan M, Ndipp RN, Shepherd AJ, et al. (2003) Evaluation of protocol using gene capture and PCR for detection ofHelicobacter pylori DNA in feaces. Journal for Clinical Microbiology 41: 4589-4593.
- Saurab KP, Chandra BP, Gopal N (2014) Diagnosis ofHelicobacter pylori; what shouldbe the gold standard? World Journal of Gastroenterology 20: 12847-12859.
- Anderson LP, Dorland A, Karankan H, Colding H, Nilsson HO, et al. (2000) Possible clinical importance of the transformation ofHelicobacter pylori intococcoid forms. Scand Journal of Gastroenterology35:897-903.
- Ndip NR, Takang MEA, Ojongokpoko AEJ, Luma HN, Malongue A, et al. (2008).Helicobacter pylori isolatesrecovered from gastric biopsies of patients with gastro-duodenal pathologies in Cameroon:status of antibiogram. Tropical Medicine and International Health 13: 848-854.
- Mégraud F, Lehours P (2007)Helicobacter pylori detection and antimicrobialsusceptibility testing. Clinical Microbiology Review 20: 280-283.