Ayush Goyal1*, Mukesh Roy2, Prabhat Gupta1, Malay Kishore Dutta1, Sarman Singh3 and Vandana Garg4
1Electronics and Communication Engg., Amity School of Engineering and Technology, Amity University, Noida, U.P., India
2Mechanical and Automation Engg., Amity School of Engineering and Technology, Amity University, Noida, U.P., India
3Clinical Microbiology & Molecular Medicine, All India Institute of Medical Sciences (AIIMS), New Delhi, India
4Internal Medicine, Max Healthcare, Noida, U.P., India
- *Corresponding Author:
- Dr. Ayush Goyal, Ph.D
Department of Electronics and Communication Engineering
Amity School of Engineering and Technology
Amity University, Noida, U.P., India
E-mail: agoyal1@amity.edu
Keywords
Mycobacterium tuberculosis; Automatic detection; Image processing; Tubeness filtering
Introduction
Tuberculosis is the leading cause of deaths due to infection worldwide. Tuberculosis (TB) detection methods include chest X-rays, culture, blood, skin, antigen, and molecular DNA tests. However, imaging of patient sputum smears is the most costeffective and widely used method for TB detection, especially in developing countries. The sensitivity of TB screening from patient sputum smears is higher when they are stained with the Ziehl- Neelsen method and imaged with fluorescent microscopy [1].
The detection of TB bacteria from sputum smears by eye is timeconsuming and prone to error as a technician or pathologist has to manually change the field of view of the microscope several times until the entire stained sputum smear on the glass slide is viewed. Automatic image processing of the stained sputum smear digital images would reduce the burden on the pathologist or technician, reduce human error, and improve sensitivity of the test. It would reduce the time required and need of lab technicians and allow preliminary detection of TB in remote or rural areas where pathologists or technicians are unavailable [2].
Identification of TB bacteria in stained sputum smear images through image processing has been performed in the past with pattern recognition techniques such as edge pixel linkage, fuzzy thresholding, and edge detection combined with morphological closing to close broken edges of segmented bacteria [3-7]. However, these methods are susceptible to error, since the stained sputum smear images have a high variability in background pixel intensity, which causes extraneous edges to be detected with these methods.
Pixel classification methods are more robust to the variation in pixel intensity in the background of Ziehl-Neelsen stained sputum smear images, and are better at detection of bacteria with unclear edges. These techniques differentiate bacteria from the background based on color, as the coating of the bacilli absorbs carbol fuchsin and is stained by its red hue whereas the background is counterstained blue by methylene [8-13].
The limitation of pixel classification approaches is that they cannot incorporate spatial or regional information about the object of interest and this regional information is necessary since each TB bacteria is a localized region or connected collection of pixels. Hence, in this research we present a method that segments the object of interest through a filtering approach. The bacteria in the stained sputum smear images have a rodlike or tubelike shape and hence they can be identified with tubeness filtering. Tubeness filtering is a type of edge or line filtering which calculates the “tubeness” or “vesselness” of each pixel in the image based on the eigenvalues of the Hessian matrix [14]. In this paper, we apply this tubeness filtering approach for the detection of mycobacterium tuberculosis in Ziehl-Neelsen stained sputum smear images.
Materials and Methods
Image Acquisition
In Ziehl-Neelsen stained sputum smear TB detection, a sputum sample is taken from patients with a cotton swab inserted into the throat. The sputum is spread on a sterile glass slide, following which it is dried and heat fixed. Then reagents are added by placing the glass slide on a staining rack with the smear side facing upward. Slowly, carbol fuschin (1%) solution is added. The slide is heated from underneath using a spirit lamp flame, until vapors start coming out. After five minutes, the slide is gently rinsed with running water. A solution of 25% sulphuric acid is added when extra water is drained off and then after 2-3 minutes standing, the slide is rinsed in running water and excess water is drained off. The process is continued by adding methylene blue solution (1%) and allowed to stand for approximately one minute. Again the slide is rinsed and after that it is ready for digital microscopic examination.
The glass slide is next inserted into a digital fluorescence microscope. The LEDs inside the microscope shine light filtered through an excitation filter to be of a particular excitation wavelength onto the glass slide and then the light reflected back from the glass slide is filtered through an emission filter and captured by a digital camera.
Image Processing
Once the digital image of the stained sputum smear is acquired, automatic image processing allows detection of TB bacteria. In the method presented herein, a special case of edge / line filtering of the image is performed. Specificially, tubeness filtering of an image gives a score to each pixel indicating how “tube-like” is the region in which the pixel falls. Literature is full of examples where tubeness filtering has been applied for detection of blood vessels or neural networks. In this paper, we apply tubeness filtering to detect rod-shaped mycobacterium tuberculosis.
The eigenvalues of the Hessian matrix are used to calculate the measure of "tubeness" [14]. If the larger two eigenvalues (λ2 and λ3) are both negative then the “vesselness” or “tubeness” for the pixel is √(λ2λ3), otherwise the value is 0. For 2D images, if the large eigenvalue is negative, the tubeness is returned as its absolute value. Else if the large eigenvalue is positive, the tubeness is calculated as zero.
The entire image is convolved with a Gaussian filter with the standard deviation (sigma) before calculating the Hessian matrix at each pixel. Specifying larger values of sigma to tune the filter select thicker tube-like structures and smaller values of sigma detect thinner tube-like structures in the image. The value of sigma depends on the radii of the tube-like structure to be extracted (in our case, 1.0 - 2.5 pixels).
Results
Figure 1 illustrates the automatic image processing of the stained sputum smear image. The RGB image is first converted to grayscale, on which tubeness filtering is applied to enhance the rod-shaped or tube-shaped bacteria. Otsu global thresholding separates the bacteria (foreground) from the background and connected component analysis labels each connected foreground region in the image as a separate object. Finally, the non-bacterial or noise regions are eliminated and only the TB bacteria remain. Finally, the number of connected regions remaining is the number of TB bacteria detected.
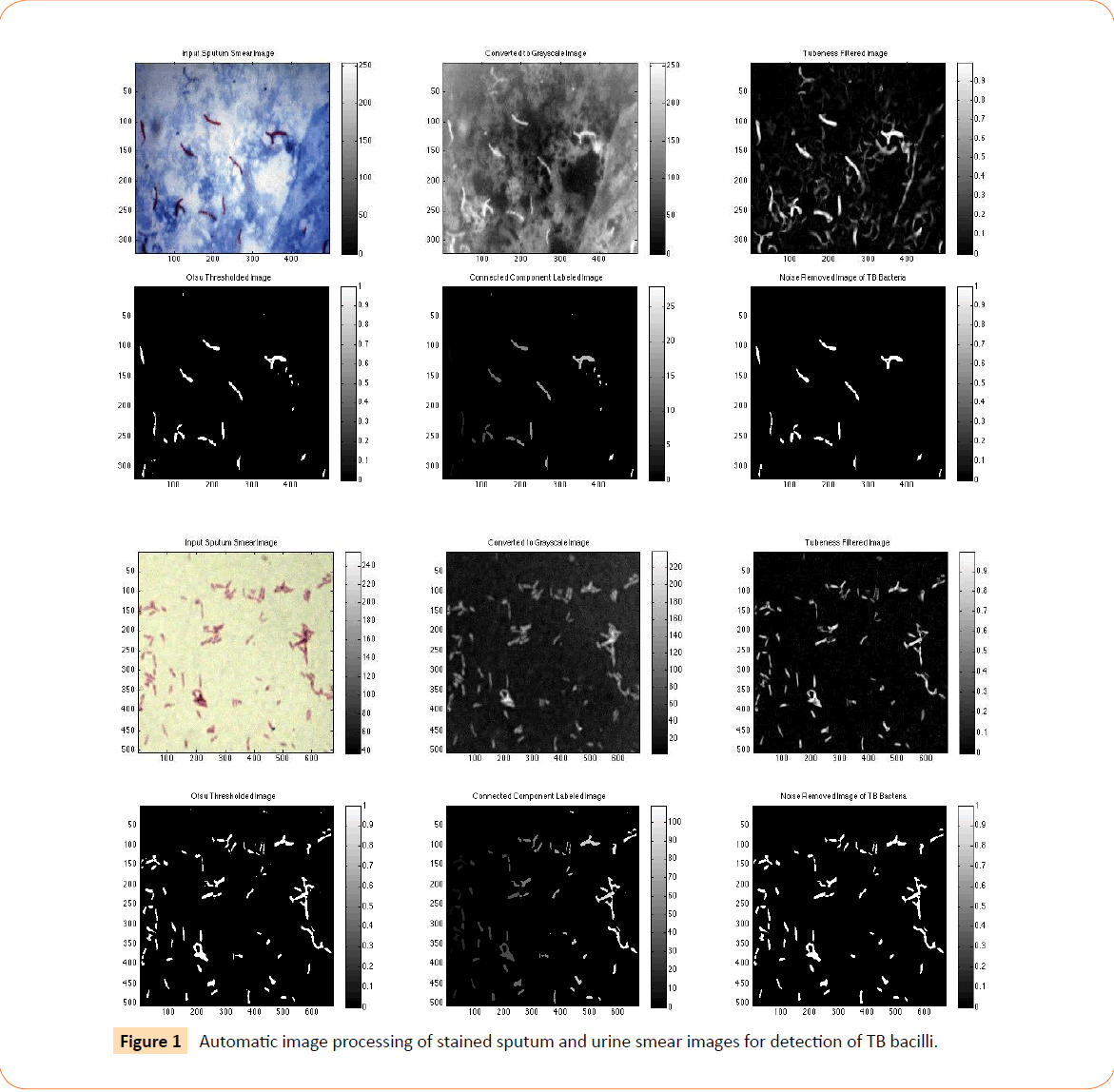
Figure 1: Automatic image processing of stained sputum and urine smear images for detection of TB bacilli.
Conclusion
The automatic detection with our algorithm presented herein results in a count of bacteria that is between the two average manual counts performed by lab technicians, and hence it is accurate for TB bacterial detection when compared to expert manual detection. Further analysis on more images with more test subjects and control images is suggested for future research to further ascertain the validity and accuracy of the automated detection of TB bacteria from stained sputum smear images with the tubeness filtering approach mentioned herein.
This research is useful for the automatic detection of mycobacterium tuberculosis from patient’s stained sputum smears. It offers two advantages in the field of tuberculosis detection – 1) Human error is eliminated since the images are automatically processed and the lab technician or pathologist does not need to manually load the glass slide into a digital fluorescence microscope and visually detect mycobacterium tuberculosis in the stained sputum smear images; 2) Quicker diagnosis of patient’s sputum smear is offered since manual detection is obviated.
This work also has future potential for determining the severity (normal, multi drug resistant - MDR, extra drug resistant - XDR, total drug resistant - TDR) of the tuberculosis based on bacterial load determined through the number of bacteria detected in the stained sputum smear image (Table 1).
| |
Expert Manual Count 1 |
Expert Manual Count 2 |
Automatic Count |
| Observation 1 |
13 |
15 |
14 |
| Observation 2 |
12 |
14 |
| Observation 3 |
14 |
15 |
| Average |
13.0 |
14.6 |
Table 1: Comparison of manual versus automatic detection of TB bacteria (each observation was performed thrice).
This research can also be extended to ascertain the type of tuberculosis a patient is affected by if stained sputum smear, stained urine smear, and stained blood smear samples are collected and imaged. If the TB bacteria are found only in the stained sputum smear image of the patient, then the patient has only pulmonary TB (PTB). However, if the TB bacteria are present in the stained blood or urinary smear images, then the patient has extra-pulmonary TB (EPTB).
6924
References
- Steingart KR, Henry M, Ng V, Hopewell PC, Ramsay A, et al. (2006) "Fluorescence versus conventional sputum smear microscopy for tuberculosis: A systematic review". Lancet Infect Dis 6: 570-581.
- Van Deun A, Salim AH, Cooreman E, Hossain MA, Rema A, et al. (2002) Optimal tuberculosis case detection by direct sputum smear microscopy: how much better is more?. Int J Tuberc Lung Dis 6: 222 -230.
- Veropoulos K, Learmonth G, Campbell C, Knight B, Simpson J (1999) Automated identification of tubercle bacilli in sputum. A preliminary investigation. Anal QuantCytolHistol 21: 277-281.
- Forero M, Sierra E, Alvarez J, Pech J, Cristobal G et al. (2001) Automatic sputum colour image segmentation for tuberculosis diagnosis. Proc SPIE 4471: 251-261.
- Forero M, Cristobal G, Alvarez-Borrego (2003) Automatic identification techniques of tuberculosis bacteria. Proc SPIE 5203: 71-81.
- Forero M, Sroubek F, Cristobal G (2004) Identification of tuberculosis bacteria based on shape and colour. Real Time Imag 10: 251-262.
- Forero MG, CristobalG, Desco M (2006) Automatic identification of Mycobacterium tuberculosis by gaussian mixture models. J Microsc 223: 120-132.
- Sadaphal P, Rao J, Comstock GW, Beg MF (2008) Image processing techniques for identifying Mycobacterium tuberculosis in Ziehl–Neelsen stains.Int J Tuberc Lung Dis 1: 579-582
- Long X, Cleveland WL, Yao YL (2004) Automatic detection of unstained viable cells in bright field images using a support vector machine with an improved training procedure. ComputBiol Med 36: 339-362.
- Meurie C, Charrier C, Lezoray O, Elmoataz A (2005) Combination of multiple pixel classifiers for microscopic image segmentation.Int J Robot Autom 20: 63-69.
- Meurie C, Lebrun G, Lezoray O, Elmoataz A (2003) A comparison of supervised pixel-based colour image segmentation methods. Application in cancerology.Proc WSEAS Trans Comput2: 739-744.
- Lenseigne B, Brodin P, Christophe T, Genovesio A (2007) Support vector machines for automatic detection of tuberculosis bacteria in confocal microscopy images. Proc 4th IEEE Symp Biomed Imag: 85-88.
- Santiago-Mozos R, Fernandez-Lorenzana R, Perez-Cruz F (2008) On the uncertainty in hypothesis testing.Proc 5th IEEE Symp BiomedImag: 1223-1226.
- Sato Y, Nakajima S, Shiraga N, Atsumi H, Yoshida S, et al. (1998) Three-dimensional multi-scale line filter for segmentation and visualization of curvilinear structures in medical images. Med Image Anal 2: 143-168.






