George T1*, Cindy B1,
Autumn G1, John B1, Ann W1,
Lina C1, Crystal T1, Rachel H1,
Virginia P1, Luebbering L1,
Meller J1, Perkins A1,
Ashley M1 and Jenell L2
1Missouri Department of Health and
Senior Services, South Jefferson St, St
Louis, MO, 63103, USA
2Department of Public Health, Saint Louis
Country
- *Corresponding Author:
- George T
Missouri Department of Health and Senior
Services, Eastern District Office, 220 South
Jefferson St, St Louis, MO, 63103, USA.
Tel: 314-877-2826
E-mail: george.turabelidze@health.mo.gov
Received date: August 08, 2017; Accepted date: August 23, 2017; Published date: August 29,
2017
Citation: George T, Cindy B, Autumn
G, John B, Ann W, et al. (2017)
Bacterial Whole Genome Sequencing in
Investigation of Salmonella Oranienburg
Infection Outbreak, Missouri, USA. Arch
Clin Microbiol. Vol. 8 No. 4:59. doi: 10.4172/1989-8436.100059
Copyright: © 2017 George T, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Keywords
Salmonella oranienburg; Outbreak, Eggs; Environmental testing; Whole genome sequencing
Introduction
In April 2015, the Missouri Department of Health and Senior Services (MDHSS) identified a cluster of four Salmonella enterica serovar Oranienburg infections among residents of the St. Louis metro area. Human cases of Salmonella serovar Oranienburg infection are relatively rare, accounting for 2.2% and 1.3% of all Salmonella serotypes in Missouri (historical surveillance data) and the United States [1], respectively. Serovar Oranienburg ranks 15th among serotypes in human salmonellosis cases in the United States. The outbreak pulsed-field gel electrophoresis (PFGE) pattern was historically rare in Missouri with only seven cases reported before 2015. In the past, serovar Oranienburg outbreaks were associated with a variety of food items, including German chocolate [2], fruit salad [3], black pepper [4], and chia powder [5]. While investigating this cluster, the MDHSS was notified by the PulseNet at the Center for Disease Control and Prevention (CDC), Atlanta, GA, USA, of the same Salmonella serovar Oranienburg isolates with the unusual PFGE pattern being reported by other states, and the multi-state outbreak investigation was initiated.
Materials and Methods
Epidemiological investigation
Epidemiological investigation was conducted by the MDHSS. Epidemiological data on the outbreak cases in other states was obtained by the MDHSS in collaboration with the respective local and state health agencies. In Missouri, a case was defined as infection with Salmonella serovar Oranienburg with the PFGE Xbal pattern JJXX01.0027 or JJXX01.1185 in a person in Missouri and the bi-state area in Illinois included in the St Louis, MO metropolitan region with illness onset occurring during April, 22-November 26, 2015. All cases were ascertained from the laboratory reports of Samonella positive specimens to the MDHSS.
A multi-state outbreak case was defined as illness in a person who was a resident of or who had known travel to Missouri and was infected with Salmonella serovar Oranienburg, with an isolate matching PFGE Xbal pattern JJXX01.0027 or JJXX01.1185, or illness in a person outside Missouri infected with Salmonella Oranienburg with isolate matching PFGE XbaI pattern JJXX01.0027 and highly related to outbreak isolates by whole genome sequencing (WGS).
Laboratory investigation
Serovar Oranienburg isolates were subtyped by the Missouri State Public Health Laboratory (MSPHL) using PFGE, and representative isolates also underwent WGS at CDC and the Food and Drug Administration (FDA). High-quality single-nucleotide polymorphism (hqSNP) analysis of human and environmental isolates was conducted by the CDC and FDA laboratories. Food samples were tested by polymerase chain reaction (PCR) at the MSPHL.
Traceback investigation
The traceback investigation was conducted by the MDHSS, and a team of MDHSS, FDA, and the US Department of Agriculture (USDA) collaborators conducted environmental investigation.
Results
A total of 46 persons meeting the case definition were identified in Missouri (44) and the bi-state area in Illinois (2). For those with known incubation period (defined as time from suspected exposure to onset of clinical illness), the median was 9 days (range: 1 to 17 days). The median age of patients was 40 years (range: <1 to 87 years); 31 (67%) patients were female. In seven (15.2%) of the 46 patients, the outbreak strain was isolated from the urinary tract (six from urine and one from renal cyst). Among 38 patients for whom information about hospitalization was available, 13 (34%) were hospitalized; one death was reported in a person with underlying medical conditions (peritoneal fluid and wound culture positive for the outbreak strain).
Initial patient interviews indicated that 25 of 46 (54%) patients reported eating at restaurant chain A during the 14 days preceding illness onset. Information from a focused questionnaire regarding food exposures at restaurant chain A revealed that 17 (68%) of the 25 patients who ate at restaurant chain A consumed eggs. Egg exposure among patients was significantly higher than among respondents of the FoodNet survey (37.8%) who reported consuming eggs away from home (p=0.002).
Eventually, a total of 52 cases were identified from 6 states, including Illinois (2), Indiana (1), Kansas (1), Missouri (44), Tennessee (3), and Texas (1) Figure 1 with illness onset dates ranging from April 1, 2015 to November 26, 2015.
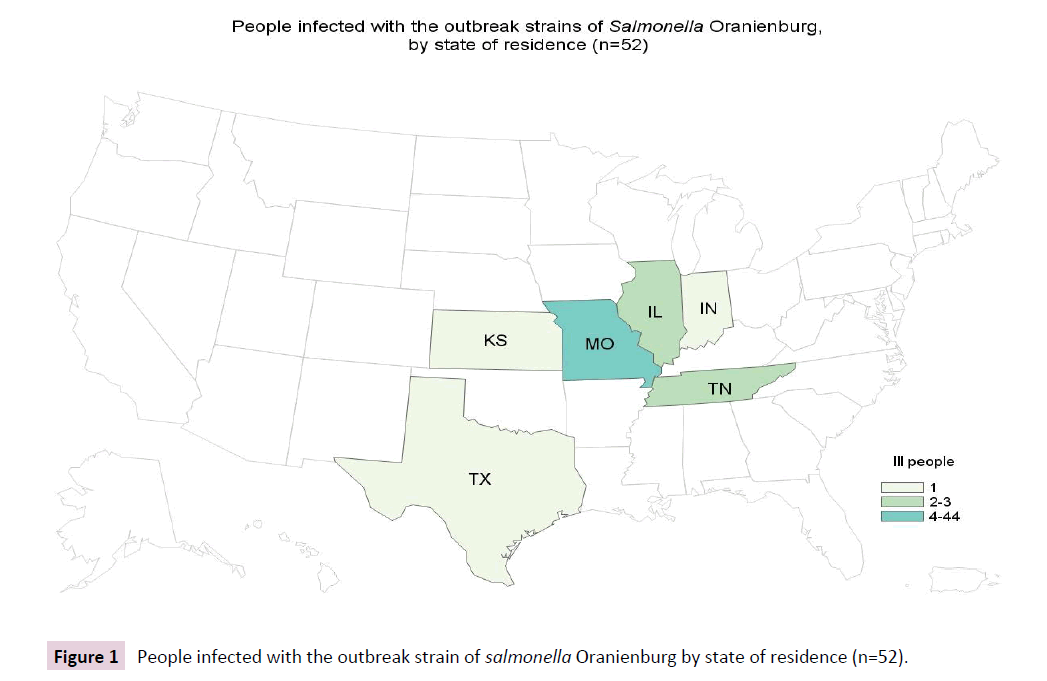
Figure 1: People infected with the outbreak strain of salmonella Oranienburg by state of residence (n=52).
All clinical isolates from Missouri were closely related by WGS using high quality single nucleotide polymorphism (SNP) analysis, but differed from seven serovar Oranienburg isolates with indistinguishable PFGE patterns reported to PulseNet from other states during the Missouri outbreak.
Food samples from one restaurant chain A location, including eggs, black pepper, margarine, and hash brown potatoes were tested by PCR at the MSPHL, and were negative for Salmonella.
The traceback investigation by the MDHSS found that all implicated restaurants used eggs from the same local egg producer. A multi-agency team conducted an inspection and environmental investigation at the implicated egg farm and egg processing facility. Through PFGE analysis, six of 30 environmental samples from the chicken farm’s laying house yielded the outbreak strain of serovar Oranienburg, and were closely related (differing by 1-7 SNPs) by WGS to the seven clinical isolates from patients with confirmed exposure to food at restaurant chain A (Figure 1, 2). Laboratory testing of the farm eggs by FDA was negative for Salmonella, but the outbreak strain was recovered from seven of 16 additional environmental specimens collected from the eggprocessing facility, including manure belts, floor samples, eggloading equipment belt, scraper, and brushes inside the dryer. WGS confirmed that the egg-processing facility environmental isolates were also closely related (differing by 1-8 SNPs) to the seven clinical isolates from patients reporting restaurant chain A exposure (Figure 1). The Xbal pattern (JJXX01.1185) of one clinical serovar Oranienburg isolate was different from the outbreak pattern by a single band, but it was highly related to the main outbreak strain based on WGS analysis. Two isolates (one environmental, one clinical) differed from the outbreak strain by the secondary enzyme BlnI by 4 bands (JJXX01.0027/ JJXA26.0029), but were also highly similar to the outbreak strain by the SNP analysis (Table 1). Isolates from patients with both clinical variants were included in the final case count.
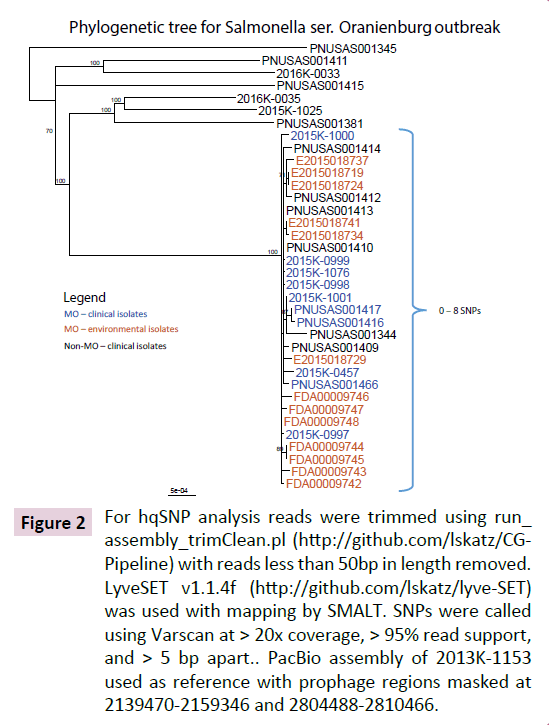
Figure 2: For hqSNP analysis reads were trimmed using run_ assembly_trimClean.pl (https://github.com/lskatz/CGPipeline) with reads less than 50bp in length removed. LyveSET v1.1.4f (https://github.com/lskatz/lyve-SET) was used with mapping by SMALT. SNPs were called using Varscan at > 20x coverage, > 95% read support, and > 5 bp apart.. PacBio assembly of 2013K-1153 used as reference with prophage regions masked at 2139470-2159346 and 2804488-2810466.
| Method (n*) |
Outbreak Case Status Change |
Total change in outbreak case status (n) |
| Yes → No |
No → Yes |
| PFGE (61) |
7 |
2 |
9 |
| WGS (16) |
0 |
0 |
0 |
*N=total tests performed on human isolates
Table 1: Change in outbreak case assignment based on combined epidemiological and laboratory data.
The six patients from Indiana (1), Kansas (1), Tennessee (3), and Texas (1) with isolates of the outbreak PFGE pattern were highly related to the Missouri outbreak by WGS. Three of these patients had known exposure in Missouri during their incubation period, and three patients were from a state that received eggs from the investigated producer (one patient reported no out-of-state travel, and two patients were lost to follow-up).
Based on the public health investigation findings, the egg farm operations were suspended, and the producer issued a voluntary recall of all eggs produced up to the time of the recall.
Discussion
This outbreak investigation identified eggs from a single poultry farm and processing facility as the likely source of the outbreak, based on the epidemiologic data, highly related Salmonella isolates from the farm and processing facility, and clinical isolates from patients. External Salmonella contamination of eggs could have occurred at either the farm or processing facility and eventually resulted in a human outbreak of salmonellosis when those eggs were consumed.
Earlier studies have shown that WGS offers increased resolution for Salmonella enterica serovar Enteritidis compared with PFGE [6]. In this investigation, application of WGS allowed health officials to exclude Salmonella isolates in other states from cases not epidemiologically linked to Missouri but with indistinguishable PFGE patterns as not related to the Missouri outbreak strain because they differed by WGS. In addition, environmental isolates from the farm and clinical isolates were shown to be highly related to one another, providing additional evidence of a link to eggs from a single producer. WGS also facilitated confirmation of the outbreak strain in patients epidemiologically linked to the outbreak, but with different PFGE patterns, and patients from other states that shared the outbreak PFGE pattern as the outbreak strain. Thus, early prospective application of WGS in this investigation was helpful for proper allocation of time and resources to investigation of cases in Missouri, rather than pursuing cases in other states that were not likely to be caused by the same source of outbreak. The WGS also allowed better data reconciliation between the laboratory findings and the epidemiological data obtained from the outbreak case interviews when compared to the PFGE analysis.
A distinguishing feature of this outbreak was an unusually prolonged incubation period (median= 9 days [range= 1-17 days]) compared with the average incubation (12-36 hours) for a nontyphoidal Salmonella. It is plausible that low-grade contamination of the eggshells resulted in a low infectious dose ingested by exposed persons, leading to the prolonged incubation period.
A similarly prolonged incubation period was reported during serovar Enteritidis outbreaks among school children in Japan, in which a low infectious dose was postulated to have resulted in a longer incubation period [7]. A second distinguishing feature of this outbreak was the high proportion of cases with urinary tract infection (UTI). The high rate of Salmonella- positive urine cultures in this study could indicate that serovar Oranienburg has an affinity for the urinary tract. An analysis of >23,000 non-typhoidal strains of Salmonella at the California public health laboratory found that 3.4% originated in urine, and that serovar Oranienburg was one of the serotypes most commonly associated with UTI [8].
Environmental contamination with Salmonella is common on poultry farms [9], and in this investigation substantial environmental contamination by Salmonella was observed on the implicated farm. In a systematic review, it was found that high level of manure contamination, middle (35-56 weeks) and late phase (>56 weeks) of the egg production cycle, high degree of egg-handling equipment contamination, flock size of >30,000, and egg production rate of >96% were risk factors associated with Salmonella contamination of shell eggs [10]; several of those risk factors were present at the farm.
One limitation of this investigation is that the prevalence of Salmonella infection in the farm flock was not estimated. It has been shown that the rate of egg shell contamination was higher in flocks with high Salmonella infection prevalence [11].
This investigation underscores importance of early prospective application of WGS in order to enhance several aspects of outbreak investigation.
20107
References
- Foodborne Diseases Active Surveillance Network (FoodNet): FoodNet Surveillance Report for (2012) Atlanta,Georgia: U.S. Department of Health and Human Services, CDC 20: 214.
- Werber D, Dreesman J, Feil F, van Treeck U, Fell G, et al. (2005) International outbreak of SalmonellaOranienburg due to German chocolate. BMC Infectious Diseases 5: 7
- Salmonella Oranienburg infections associated with fruit salad served in health-care facilities--northeastern United States and Canada, 2006 (2007) MMWR Morb Mortal Wkly Rep 56: 1025-1028.
- Gustavsen S, Breen O (1984) Investigation of an outbreak of Salmonella oranienburg infections in Norway, caused by contaminated black pepper. Am J Epidemiol 119: 806-812.
- Multistate Outbreak of Salmonella Infections Linked to Organic Sprouted Chia Powder (2014) Am J Epidemiol 152.
- Deng X, Shariat N, Driebe E, Roe CC, Tolar B, et al. (2015) Comparative analysis of subtyping methods against a whole-genome-sequencing standard for Salmonella enterica serotype Enteritidis. J Clin Microbiol 53: 212-218
- Abe K, Saito N, Kasuga F, Yamamoto S (2004) Prolonged incubation period of salmonellosis associated with low bacterial doses. J Food Prot 67:2735-2740.
- Abbott SL, Portoni BA, Janda JM (1999) Urinary tract infections associated with nontyphoidal Salmonella serogroups. J Clin Microbiol. 37: 4177–4178.
- Berghaus RD, Thayer SG, Law BF (2013) Enumeration of Salmonella and Campylobacter spp. in Environmental Farm Samples and Processing Plant Carcass Rinses from Commercial Broiler Chicken Flocks. Applied and Environmental Microbiology 79: 4106-4114.
- Denagamage T, Jayarao B, Patterson P, Wallner-Pendleton E, Kariyawasam S (2015) Risk Factors Associated With Salmonella in Laying Hen Farms: Systematic Review of Observational Studies. Avian Dis 59: 291-302.
- Arnold ME, Martelli F, McLaren I, Davies RH (2014) Estimation of the Rate of Egg Contamination from Salmonella-Infected Chickens. Zoonoses Public Health 61:18-27.







