Keywords
Enterobacteriaceae; Quinolone resistance determinants; aac(6’)Ib-cr
Introduction
Development of bacterial resistance towards various classes of antibiotics and spread of resistance genes depends on the situation that is initiated by over-the-counter availability, indiscriminate and inappropriate use of antimicrobial agents especially in a country like India [1-3]. An alarming situation is the progressive loss of susceptibility towards ciprofloxacin and norfloxacin due to its increased use in the treatment of a broad range of clinical conditions like urinary tract infections, upper respiratory tract infections, and as a prophylaxis in neutropenic patients, as well as in the poultry sectors [4].
Antibiotic resistance is particularly predominant among Gram -negative bacteria, specifically within the members of Enterobacteriaceae [5-7]. Furthermore, fluoroquinolone resistant bacterial isolates are disseminating in the environment through hospital wastes and if not treated properly may find their way into water bodies.
Antibiotics used in medicine are only partially metabolized [8] by patients, and discharged into the hospital sewage system or directly into the municipal waste- water. The predicted concentrations of antibiotics in hospital wastewater are in the range of the sub- inhibitory concentration (MIC50) of sensitive pathogenic or beneficiary bacteria for some active substances (0.1 - 2.9 mg/l) leading to the growing antibiotic resistance within the bacterial flora led to lateral transfer of antibiotic resistance[9] among pathogens as well as in non-pathogenic microorganisms thereby making them reservoirs for maintenance, propagation and expansion of resistance genes. Quinolones, to be the most commonly used antibiotic in community acquired infections, have become ineffective due to increasing resistance in recent years. However, no such study from this country is available to present a scenario of quinolone resistance and their maintenance within environmental enterobacterial isolates. A study representing the presence of quinolone resistant isolates and their molecular basis would be of epidemiological and therapeutic importance.
Thus, the present study was undertaken to investigate the occurrence of quinolone resistance determinants within environmental and food isolates and their transmission dynamics.
Materials and Method
Sample were collected from river water (Barak river, ghagra river, longai river, singla river, jatinga river) from five different sites of each river (n=25). Water samples were also collected from other bodies (n=11) near waste disposal while ready-toeat food samples (n=48) were also collected from food vendors shop Samples were collected from March 2015 to August 2015. All the samples were microbially processed using appropriate techniques for isolation of Enterobacteriaceae, while isolates were further confirmed by microscopical investigation, cultural characteristics and standard biochemical reactions [10].
Screening of quinolone resistance
Using the disc diffusion method of the CLSI recommendations, quinolone resistance was determined on the following antibiotics Nalidixic acid (30 μg), Norfloxacin (10 μg), Ciprofloxacin (5 μg), Ofloxacin (5 μg), Lomefloxacin (5 μg), Levofloxacin (5 μg), (Himedia, Mumbai, India). E.coli ATCC 25922 served as control for antimicrobial susceptibility tests. Minimum Inhibitory Concentration (MIC) of isolates towards norfloxacin (Norflox, Cipla Ltd, Sikkim), ciprofloxacin (Ciplox, Cipla Ltd, Sikkim), ofloxacin (Oflacin, Micro Labs Ltd, Bangalore) and levofloxacin (Levotop-PF 1.5%, Ajanta Pharma Limited, Mumbai) was also determined by agar dilution method and results were interpreted as per CLSI guidelines [11].
Characterization of quinolone resistance by multiplex PCR assay
DNA extraction was performed using an improved boiling centrifugation method [12]. Presence of qnrA, qnrB, qnrS, qnrD, qnrC and aac (6’)-Ib-cr genes was detected by PCR based technique using the primers shown in Table 1 by thermal cycler (Bio-Rad, USA). Each single reaction mixture (25 μl) contained 1 μl ( 10 ng) of DNA suspension, 15 pmol of each primer, 12.5 μl of 2x Go green master mix (Promega,Madison, USA) and nuclease free water is added to make the volume 25 μl. Previously screened qnrA, qnrB, qnrS, qnrD, qnrC and aac (6’)-Ib-cr positive isolates was taken as positive control and E.coli ATCC 25922 was taken as negative control. Reactions were run under the following conditions; initial denaturation at 95°C for 2 min; 35 cycles of 95°C for 50 sec, 53°C for 40 sec and 72°C for 1.20 min; and a final extension at 72°C for 5 min.
| Primer pair |
Target |
Sequence (5'
3') |
Product size
(bp) |
Reference |
qnrA-1A
qnrA-1B |
qnrA |
TTCAGCAAGAGGATTTCTCA
GGCAGCACTATTACTCCCAA |
628 |
23 |
qnrB-CS-1A
qnrB-CS-1B |
qnrB |
CCTGAGCGGCACTGAATTTAT
GTTT
CTGCTCGCCAGTCGA |
546 |
23 |
qnrS-1A
qnrS-1B |
qnrS |
CAATCATACATATCGGCACC
TCAGGATAAACAACAATACCC |
675 |
23 |
qnrC-F
qnrC-R |
qnrC |
GGGTTGTACATTTATTGAATC
TCCACTTTACGAGGTTCT |
447 |
22 |
qnrD-F
qnrD-R |
qnrD |
CGAGATCAATTTACGGGGAATA
AACAAGCTGAAGCGCCTG |
582 |
19 |
aac(6_)-Ib-cr-F
aac(6_)-Ib-cr-R |
aac(6_)-Ib
cr |
ATG ACT GAG CAT GAC CTT GC
TTA GGC ATC ACT GCG TGT TC |
519 |
22 |
Table 1: List of primers.
Plasmid analysis and transformation
Plasmid DNA was extracted and purified by Qiagen mini prep kit (Germany). The plasmid was transferred into E.coli DH5? by the heat shock method [13] and transformants were selected by incubation on Luria– Bertani (LB) (Himedia, Mumbai, India) agar plates containing 0.25 μg/ml and 0.5 μg/ml of norfloxacin, ciprofloxacin and levofloxacin each. Transformants were screened for their plasmid content and resistance phenotype. To investigate the transferability of plasmid encoding quinolone resistance, conjugation experiment was performed using the streptomycinresistant E.coli recipient strain B (Genei, Bangalore, India) were performed as described previously [14].
Results
A total of 68 collected isolates were identified as Enterobacteriaceae which were Escherichia coli (n = 23), Klebsiella pneumoniae (n =4), Klebsiella oxytoca (n =22), Proteus mirabilis (n=19). Fifty seven among them were found to be resistant to at least one of the quinolone group of drug tested (Table 2). Common Resistance profiles were nalidixic acid (89.17%) followed by norfloxacin (81.49%) and ciprofloxacin (73.56%). Results of Minimum Inhibitory Concentration were shown in Table 3.
| OrganismAntibiotics |
Escherichia coli
(N=23) n% |
Klebsiellapneumoneae
(N=4) n % |
Klebsiellaoxytoca(N=22) n % |
Proteus mirabilis
(N=19) n % |
| Nalidixic acid |
19 |
82.61 |
3 |
_ |
21 |
95.45 |
17 |
89.47 |
| Norfloxacin |
17 |
73.91 |
3 |
_ |
19 |
86.36 |
16 |
84.21 |
| Ofloxacin |
14 |
60.87 |
2 |
_ |
17 |
77.27 |
13 |
68.42 |
| Ciprofloxacin |
15 |
65.21 |
2 |
_ |
18 |
81.81 |
14 |
73.68 |
| Lomefloxacin |
14 |
60.87 |
3 |
_ |
17 |
77.27 |
14 |
73.68 |
| Levofloxacin |
13 |
56.52 |
1 |
|
14 |
63.6 |
10 |
52.63 |
Table 2: Antibiogram profiling of quinolone antibiotics.
| Isolates |
MIC range of quinolone resistant isolate(µg/ml) |
| NX |
CIP |
OF |
LEV |
| E.Coli |
64 - ≥128 |
32- ≥64 |
16- 64 |
16- 32 |
| Klebsiellapneumoneae |
4- ≥64 |
8- 128 |
8-≥64 |
16- 32 |
| Klebsiellaoxytoca |
32- 64 |
64 |
16-64 |
8 |
| Proteus mirabilis |
128 |
64-128 |
32 |
8 |
NX- Norfloxacin, CIP- Ciprofloxacin, OF- Ofloxacin, MIC-Minimum Inhibitory Concentration.
Table 3: MIC of quinolone resistant isolate.
Based on the PCR results, the prevalence of aac(6’)Ib-cr was highest (n=23) followed by qnrD (n=7), qnrA (n=5) and qnrS (n=2). None of the isolates showed the presence of qnrC (Table 4) and (Figures 1 and 2).
| Qnr determinants |
Total no. of isolates |
| qnrA |
5 |
| `qnrD |
7 |
| qnrS |
2 |
| Aac(6’)Ib-cr |
23 |
Table 4: Distribution of qnr determinants.
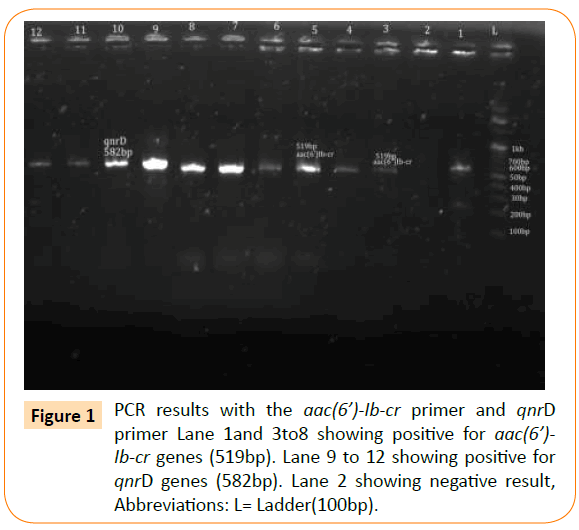
Figure 1: PCR results with the aac(6’)-Ib-cr primer and qnrD primer Lane 1and 3to8 showing positive for aac(6’)- Ib-cr genes (519bp). Lane 9 to 12 showing positive for qnrD genes (582bp). Lane 2 showing negative result, Abbreviations: L= Ladder(100bp).
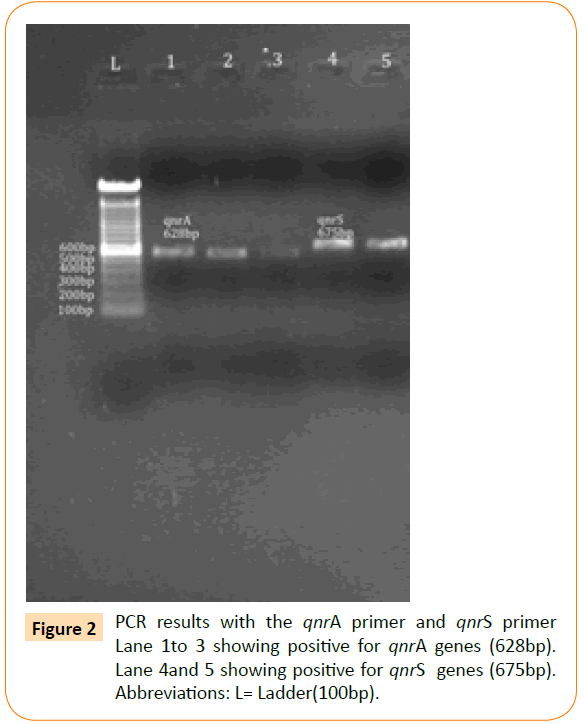
Figure 2: PCR results with the qnrA primer and qnrS primer Lane 1to 3 showing positive for qnrA genes (628bp). Lane 4and 5 showing positive for qnrS genes (675bp). Abbreviations: L= Ladder(100bp).
Plasmid DNA encoding qnrS, aac(6’)Ib-cr, qnrA and qnrD were transformed in E.coli DH5? in media containing ciprofloxacin, norfloxacin and levofloxacin. Only qnrS and aac(6’)Ib-cr were able to get transformed. The transformants carrying qnrS gene was selected from the media containing ciprofloxacin and the transformants carrying aac(6’)Ib-cr gene were selected from the media containing both norfloxacin and ciprofloxacin. The transformants carrying qnrS and aac(6’)Ib-cr genes were confirmed by PCR. To check the self-transferability, the isolates carrying the qnrS and aac(6’)Ib-cr were subjected to conjugation using the streptomycin-resistant Escherichia Coli recipient strain B. The trans-conjugants were selected in media containing streptomycin and ciprofloxacin. Both of the genes could be horizontally transferred in E.coli recipient strain B by conjugation whereas other types i.e., qnrA and qnrD could not be conjugatively transferred.
Discussion
Fluroquinolone resistant enterobacteria isolated from water bodies, waste disposal and food sample, whether pathogenic or not, may come into contact with other microbes and transfer resistant genes. In this study species most prevalent gut colonizer E.coli, comprised 33.82% of total enterobacterial isolates in tested samples, which is an indicator of faecal contamination. This is in accordance with the study carried out by Chen et al. in China [15] where the prevalence of E.coli was 36.4%.In another study carried out to assess the microbiological quality of ready to eat street vended food in porto region of potugal [16] where E.coli accounts for 55%. The presence of E.coli in food sample indicates the lateral entry of enterobacterial isolates into food chain which may cause infection. Compared to this study, another report showed a different distribution in Mexico city [17]. Many studies were performed on the phenotypic detection of PMQR genes by using mostly nalidixic acid as an indicator [18]. The study carried out by Cavaco et al. [19] suggested that nalidixic acid alone did not confer the maximum effectiveness for the detection of these resistance determinants and used both antibiotics for the detection of PMQR, which is almost similar to our study where 39% of the isolates were simultaneously resistant to norfloxacin, ciprofloxacin and ofloxacin. The most prevalent were aac(6’)Ib-cr followed by qnrD, qnrA and qnrS. This is in contrast with the observation made by Marti et al. [20] and Poirel et al. [21-23] which stated that qnrS was the most commonly identified acquired qnr gene in the environment, because it is usually identified in waterborne species. Transferability of the qnrS and aac(6’)-Ib-cr gene in this study suggests that there is a horizontal transmission of these genes among the environmental isolates.
Thus comparing with all the previous reports, our study showed different pattern with diverse quinolone resistance determinants were carried within environmental isolates. Maintenance of diverse resistant genes in the environment could be due to irrational use of quinolone group of antibiotics in the community where these drugs are available over the counter.
Conclusion
Remarkable rates of colonisation with high-level fluroquinolone resistance were reported among multidrug resistant community Enterobacterial isolates in the current study which also highlighted the presence of these resistant determinants within environmental isolates.
Acquisition of resistance genes could be a natural process in the microorganism and the competing environment helps their maintenance in subsequent generations. However, the main concern in this phenomenon is how this gene transfers and their persistence affects the clinical settings and treatment alternatives. This kind of study not only identifies the resistance determinants but also predict their mobilization, source of acquisition, origin and evolution. As quinolone remains mostly prescribed oral antibiotics, the resistance against this group especially in community infection leads into severe clinical implications. Thus, the present study can be concluded that the presence of these plasmid mediated quinolone resistance determinants in environmental isolates particularly aac(6/)Ib-cr is indicative of its ability of persistence ,thus causing significant health hazard. Also the source of these genes could be from normal gut flora which is acquired during infection followed by a course of quinolone therapy, released into the environment through faecal contamination or vice versa.
Proper measures should be adopted in public health and hygiene management. So as the trace and prevent the expansion of this health hazard in community environment.
Financial Support
Department of Bio- technology (DBT-NER, Twinning) Government of India
Conflict of Interest
None to declare
Acknowledgment
The authors would like to acknowledge the help of Dr. Piyush Pandey, HOD Microbiology, Assam University for providing infrastructure as well as the financial support provided by Department of Biotechnology (DBT-NER twinning Scheme) to carry this study. Authors also acknowledge the Assam University Biotech Hub for providing laboratory facility to complete this work.
7472
References
- Okeke IN, Lamikanra A, Edelman R (1999) Socio-economic and behavioral factors leading to acquired bacterial resistance to antibiotics in developing countries. Emerging Infectious Diseases 5: 18-27.
- Sahm DF,Karlowsky JA, Kelly LJ,Critchley IA, Jones ME, et al. (2001) Need for annual surveillance of antimicrobial resistance in Streptococcus pneumoniae in the United States: 2-year longitudinal analysis. Antimicrobial Agents and Chemotherapy 45: 1037-1042.
- Lockhart SR, Abramson MA, Susan EB, Gallagher G, Riedel S, et al. (2007) Antimicrobial resistance among Gram-negative bacilli causing infections in inten-sive care unit patients in the United States between 1993 and 2004. Journal of Clinical Microbiology 45: 3352- 3359.
- Bazile-Pham-Khac S, Truong QC, Lafont JP, Gutmann L, Zhou XY, et al. (1996) Resistance to fluoroquinolones in Escherichia coli isolated from poultry. See comment in PubMed Commons below Antimicrob Agents Chemother 40: 1504-1507.
- Thomson CJ (1999) The global epidemiology of resistance to ciprofloxacin and the changing nature of antibiotic resistance: A 10-year perspective. Journal of Antim- icrobial Chemotherapy, 43: 31-40.
- Toltzis P (2004) Antibiotic-resistant gram-negative bacteria in hospitalized children. See comment in PubMed Commons below Clin Lab Med 24: 363-380.
- Schlackow I, Stoesser N, Walker AS, Crook DW, Pefo TEA, Wyllie DH (2012) Increasing incidence of Escherichia coli bacteraemia is driven by an increase in an- tibiotic-resistant isolates: Electronic database study in Oxfordshire 1999-2011. Journal of Antimicrobial Chemotherapy, 67: 1514-1524.
- Martinez JL (2009) Environmental pollution by antibiotics and by antibiotic resistance determinants. See comment in PubMed Commons below Environ Pollut 157: 2893-2902.
- Kümmerer K (2001) Drugs in the environment: emission of drugs, diagnostic aids and disinfectants into wastewater by hospitals in relation to other sources--a review. See comment in PubMed Commons below Chemosphere 45: 957-969.
- Holt JG, Krieg NR, Sneath PHA, Staley JT, Williams ST (2000).Bergey’s manual of Determinative Bacteriology; ninth edition 203-222.
- Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing: twenty-first informational supplement M100-S21. CLSI. 2012
- Cattoir V, Poirel L, Rotimi V, Soussy CJ, Nordmann P (2007) Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. See comment in PubMed Commons below J AntimicrobChemother 60: 394-397.
- Li S, Anderson ML, Yang H (2007) Applied Physics Letters 91, 013902 Heat shock method.
- Oktem IM, Gulay Z, Bicmen M, Gur D; HITIT Project Study Group (2008) qnrA prevalence in extended-spectrum beta-lactamase-positive Enterobacteriaceae isolates from Turkey. See comment in PubMed Commons below Jpn J Infect Dis 61: 13-17.
- Chen X, Zhang W, Pan W, Yin J, Pan Z, et al. (2012) Prevalence of qnr, aac(6')-Ib-cr, qepA, and oqxAB in Escherichia coli isolates from humans, animals, and the environment. See comment in PubMed Commons below Antimicrob Agents Chemother 56: 3423-3427.
- Campos J, Gil J, Mourao J, Peixe L, Antunes P (2015) Ready to eat street- vended food as a potential vehicle of bacterial pathogens and antimicrobial resistance : An exploratory study in Porto region, Portugal. International Journal of Food Microbiology 206:1-6.
- Amábile-Cuevas CF, Arredondo-García JL, Cruz A, Rosas I (2010) Fluoroquinolone resistance in clinical and environmental isolates of Escherichia coli in Mexico City. See comment in PubMed Commons below J ApplMicrobiol 108: 158-162.
- Mirarini LAR, PoirelL,Cattaoir V, Darini ALC,Nordmann P (2008). Plasmid- mediated quinolone resistance determinants among enterobacterial isolates from outpatient in Brazil. Journal of antimicrobial chemotherapy 62: 474-478.
- Cavaco LM, Hasman H, Xia S,Aarestrup FM (2009) qnrD, a novel gene conferring transferable quinolone resistance in Salmonella entericaserovars Kentucky and Bovismorbificans of human origin. Antimicrob. Agents Chemother 53:603–608.
- Marti E, Balcázar JL (2013) Real-Time PCR assays for quantification of qnr genes in environmental water samples and chicken feces. See comment in PubMed Commons below Appl Environ Microbiol 79: 1743-1745.
- Poirel L, Cattoir V, Nordmann P (2012) Plasmid-Mediated Quinolone Resistance; Interactions between Human, Animal, and Environmental Ecologies. See comment in PubMed Commons below Front Microbiol 3: 24.
- Wang M, Guo Q, Xu X, Wang X, Ye X, et al. (2009) New plasmid-mediated quinolone resistance gene, qnrC, found in a clinical isolate of Proteus mirabilis. See comment in PubMed Commons below Antimicrob Agents Chemother 53: 1892-1897.
- Cano ME, Rodriguez-Martinez JM, Aguero J, Pascual A, Garcia-Lobo JM, et al. (2006) Detection of qnrS in clinical isolates of Enterobacter cloacae in Spain, 16thEuropean Congress of Clinical Microbiology and Infectious Diseases; Nice, France; April 1 Abstract O52.







