Keywords
Enzyme regulation; Drug; Classification
Introduction
The exposition of working mechanism of enzyme regulators is of great importance for both enzymology and pharmacology. In vivo, some drugs take their physiological role by acting regulator, activator or inhibitor, of enzyme, receptor, and protein. However, the current view about working mechanism of enzyme regulator, or allosteric regulation mechanism, is experimental and a comprehensive understanding of it cannot be obtained [1-3]. The key theoretical flaws of allosteric regulation are: 1) it is experimental model and theoretical foundation of enzyme regulation cannot be resolved within it; 2) it can only explain effect of some, but not all factors and substances involved in enzyme regulation; 3) the integration mechanism of many types of regulators within enzyme molecule cannot be explained by this model; 4) the logic relation between protein thermodynamics or dynamics and enzyme regulation has been ignored; 5) the quantitative description of enzyme regulation cannot be done.
In order to resolve such problems, two approaches have been processed. One stresses the role of protein dynamics in enzyme regulation [4,5]. However, the relationship among protein dynamics, protein thermodynamics and protein conformation, cannot be resolved within landscape theory of protein dynamics and this impedes the first approach to be developed into a more matured theory of enzyme regulation. In another way, we have proposed thermodynamic model for enzyme regulation on the basis of protein thermodynamics and protein thermodynamic structure theory. According to protein thermodynamic structure theory, any protein conformational state in terms of biophysics, or conformation in terms of biochemistry, represents an ensemble of protein conformation at atom level. With this cognition, most views of dynamic model of allostery can be accepted within our model [6-9].
This paper will discuss theoretical classification of enzyme regulator on the basis of thermodynamic model for enzyme regulation [8,9]. Within our model, enzyme regulator refers all factors and substances, physical and chemical, inhibitor or activator, which can affect enzyme activity. The salt concentration, protein phosphorylation, gene mutation, temperature, pH of working buffer represent different regulators of enzyme activity. In contrast, most experimental scientists do not consider these factors in their study of enzyme regulation.
According to our model, a regulator of enzyme can influence enzyme activity by two different ways. By first way, a regulator, or allodynamic regulator of enzyme, influence abundance of active conformation of enzyme in solution and thus adjust enzyme activity. By second way, regulator, or local modulator of enzyme, affect subtle thermal state of conformation of enzyme at active site, and then alter properties of enzymatic reaction. Thus, enzyme regulators can be grouped according to their working mechanism.
Ideal Allodynamic Regulator of Enzyme Activity
In the first mechanism of enzyme regulation, the abundance of active conformation of enzyme can be regulated by regulatory conformations of enzyme. A regulator, which binds to regulatory conformation of enzyme, will change thermal state of an enzyme and influence thermal equilibrium between active and inactive conformation of enzymes. If regulator binds to an area of enzyme which is far away from active site of enzyme and there is no direct interaction between regulatory site and active site of enzyme, all interaction between them are carried out by global conformation of enzyme. This type of enzyme regulation is called allodynamic.
In order to get comprehensive understanding of its working mechanism, we need to examine many conformations involved in enzyme regulation. According to protein thermodynamic structure theory, a protein shows numerous conformations which can be theoretically isolated or labeled by a thermodynamic parameter or function under different conditions. For an enzyme, the enzyme activity is logically related to the conformation at active site (CAS), most parts of the enzyme have no direct relation to the execution of enzyme activity, but has ability to regulate enzyme activity by adjusting thermodynamic state of CAS via some ways. The regulatory conformation of the enzyme (RCE) refers to a part or site of the enzyme which can bind a regulator or respond environmental factors. The active conformation of the enzyme (ACE) represents global conformation which control the local conformation (CAS or RCE). The RCE and ACE share some common components and thus the thermal state of two conformations can be interacted or coupled with each other. In some oligomeric enzymes, the regulatory subunit represents a specific case for RCE [10].
The thermodynamic state of RCE can be judged by a change of binding affinity between regulator and enzyme and the potency of regulator in enzyme regulation. The state of CAS can be judged by a change of parameters of enzymatic reaction, such as maximum rate (Vm), km, and activation energy. The active or inactive formation of enzyme has been well defined in enzymology. The difference between active conformation and CAS has also been well defined [11].
The protein thermodynamic structure of enzyme regulator complex is shown in figure 1.
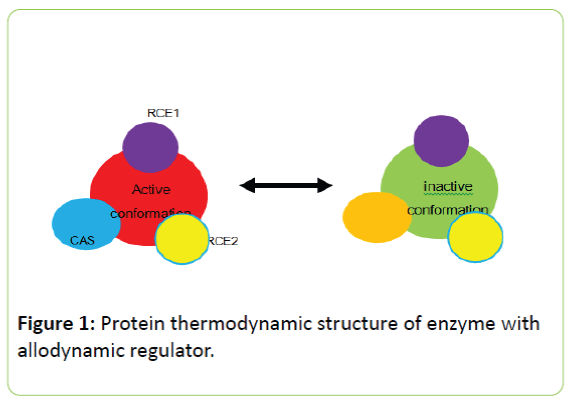
Figure 1: Protein thermodynamic structure of enzyme with allodynamic regulator.
An enzyme has many types of regulatory conformation and they are located at different sites of the enzyme, but two are pictured in this figure. The active (or inactive) conformation of enzyme is at global level and it controls local conformations (substrate binding conformation and regulatory conformation). Two conformations, such as ACS and active conformation, may share same components and therefore they are overlapped.
When binding to a regulator or respond to a stimulus of environment, the thermal state of RCE firstly changes. The change of RCE can then influence the thermal state and probability of active conformation of enzyme. The thermal change of ACE can further induce a change of conformational state at active site of enzyme. Thus, the thermal change at RCE can be transmitted into CAS. This processed is called reorganization of protein thermodynamic structure. According to our model, the allosteric regulation represents one specific fashion for allodynamic regulation.
The information transmission way of allodynamic regulation of an enzyme can be expressed as follows:
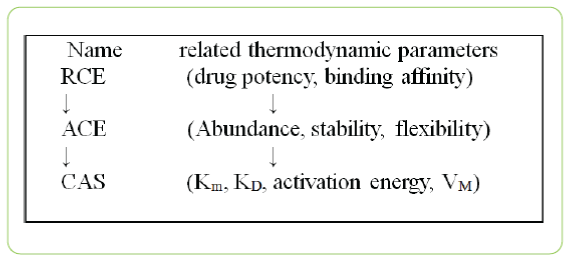
Although the thermodynamic state of protein conformation involved in enzyme regulation cannot be measured in enzymology, we can define and quantify the pathway of allodynamic regulation by diversified parameters of enzymatic reaction and enzyme regulation. The co-change or the relation among different properties of enzyme regulation or parameters of enzymatic reaction can be clearly seen in active conformation curve of enzyme [8]. The change of abundance of active conformation of enzyme confirms that that enzyme regulation mechanism can be only fully explained by protein thermodynamics for it can be quantitatively and precisely described by laws of thermodynamics. The change of protein dynamics or flexibility is precondition for enzyme regulation, but not sufficient condition. Or in other words, the matured theory for enzyme regulation cannot be established on the basis of protein dynamics.
By applying principles of protein thermodynamic structure theory, following properties and criteria of the ideal allodynamic regulator of an enzyme could be obtained and it could be utilized to judge the nature of enzyme regulator.
There is no direct physical interaction between substrate binding conformation and regulator binding conformation. Their interaction is totally indirect, or via active conformation of enzyme.
Although there is indirect interaction between regulator binding conformation and substrate binding conformation, the binding affinity between enzyme and substrate is not influenced by regulator. Two underlying reason are: 1) enzyme substrate complex (or regulator enzyme complex) can show a large number of nearly isoenergetic conformations, energy involved in protein conformational change is rather small [12]; 2) any protein conformation shows independence in some degree [6]. Thus, the indirect interaction between RCE and ACS may not be expressed in binding ability between substrate and enzyme. Similar opinion in pharmacology is that the binding between receptor and agonist may be not influenced by regulator.
In the presence of allodynamic regulator, at least one parameter of enzymatic reaction (maximum rate (Vm), the km, and activation energy of enzymatic reaction) will be changed. But unlike to allosteric model, the change of km is not crucial.
The nature of enzyme regulation is same for all substrates if an enzyme has many substrates. It is crucial evidence for this type of regulation. This property has not been studied experimentally. This property can be judged by many criteria: a) if a regulator shows inhibition to enzyme activity for one substrate, it will show inhibitory activity when different substrates of enzyme are used; b) if a regulator increases the km for one substrate of enzyme, it will increase the km of other substrate by same way; and etc.
The efficiency of regulator is changeable under different conditions. The inhibitor may become activator under different condition. The allodynamic regulator (inhibitor) of enzyme may lose its effect if the protein flexibility is changed. In receptor field, it is called receptor desensitization.
Although noncompetitive inhibitor of enzymatic reaction was judged by different criterion in experimental science, within our model, they are ideal allodynamic regulator [11].
According to our model, the so called allosteric regulation of hemoglobin is perfectly carried out by allodynamic regulation mechanism. In this case, one molecule of oxygen acts regulator for the binding between hemoglobin and other oxygen molecules. The pH (or H+) act regulator for allodynamic regulation and it was called Bohr effect [11]. It is example of the effect integration among different regulators.
Local Modulator of Active Site Conformation of Enzyme (Biased Regulator)
For second mechanism of enzyme regulation, the fine conformational state of enzyme at active site is directly modulated by regulator. If RCE is at adjacent area of ACS, there is direct communication between them, the RCE can influence thermal state of ACS directly and this regulation has no relation to global conformation of enzyme or CAS. The protein thermodynamic structure of this type of enzyme regulator complex is shown in figure 2.
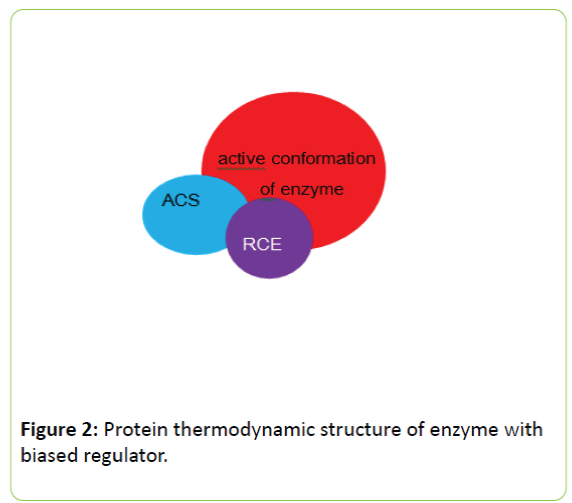
Figure 2: Protein thermodynamic structure of enzyme with biased regulator.
In this fashion of enzyme regulation, the regulator binds to an area near active site of enzyme. The binding of regulator alters the fine conformation of active site of enzyme. Within in this fashion of enzyme regulation, the global conformational state of enzyme may not be influenced by regulator.
By applying principles of protein thermodynamic structure theory, properties of this type regulation can be deduced as follows:
There is direct interaction (or overlapping) between regulator binding conformation and substrate binding conformation, and this type interaction is independent to global conformation of enzyme.
The activation energy, maximum rate, and km of enzymatic reaction may be all changed.
The property of this type regulation is changeable from substrate to substrate. In other words, it is biased regulator [13].
A good example of this type of regulation is ribonucleotide reductase. For this enzyme, dATP, TTP, and dGTP act local modulator of enzyme activity [14].
Within this model, the competitive inhibition could be considered a special case for this type of enzyme regulation. It is possible to find a substance which acts competitive inhibitor for a substrate of enzymatic reaction, and it acts local modulator for another substrate.
The mechanisms above mentioned for enzyme regulator represent two typical cases. For a substance or drug, its effect on enzymatic reaction may be complex and could be carried out by two mechanisms at same time.
Effect Integration Among Different Regulators
A highlighted advantage of our model is that integration of effects of different regulators on enzyme activity can be easily explained. All types of information of regulators is integrated thermodynamically within enzyme molecule and this feature plays important role in signaling activity of enzymes [15]. For first mechanism of enzyme regulation, the effect of different regulators on enzyme activity can be freely integrated for they all influence the abundance of active conformation of enzyme. The cooperation and neutralization of effect of regulators represent two extremely cases for effect integration. The shift of active conformation curve along an environmental factor represents third type of effect integration of different factors or regulator on enzyme activity [8]. When two local regulators of enzyme are integrated, the case will be complex and there is no general rule about it.
Finally, differences and consensus of two type regulator is summarized in Table 1.
| |
Ideal allodynamic regulator |
Biased regulator |
| Regulatory conformation of enzyme |
Yes |
UI |
| Km, KD, kcat, Ea of enzymatic reaction |
UI |
Yes |
| Similarity for different substrates |
Yes |
No |
| Far away from active site |
Yes |
No |
| Sensitivity to environment |
high |
Low |
| Cooperation among same group |
Yes and free |
NR |
| Regulatory sites |
Many |
Few |
KD: binding constant. UI: unnecessary to influence all these parameters. NR: there is no rule about it.
Table 1: Summary of nature of ideal allodynamic regulator and local modulator of enzyme activity.
Conclusion
In conclusion, the enzyme regulation mechanism and working mechanism of drug can be fully explained by applying principles of protein thermodynamics and protein thermodynamic structure theory. In order to get comprehensive understanding of working mechanism of enzyme regulator and drug, the active conformation curve of enzymes along many variables should be measured experimentally in future.
10600
References
- Koshland DE, Némethy G, Filmer D (1966) Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry 5: 365-385.
- Monod J, Wyman J, Changeux JP (1965). On the nature of allosteric transitions: a plausible model. J MolBiol 12: 88-118.
- Changeux, JP (2012). Allostery and the Monod-Wyman-Changeux model after 50 years. Annual review of biophysics 41: 103-133.
- Kern D, Zuiderweg ER (2003) The role of dynamics in allosteric regulation. CurrOpinStructBiol 13: 748-757.
- Motlagh HN, Wrabl JO, Li J, Hilser VJ (2014). The ensemble nature of allostery. Nature 508: 331-339.
- Zhao Q (2012) Partition function of protein conformational state. J ComputTheorNanosci9: 745-751
- Zhao Q (2015) A thermodynamic and theoretical view for enzyme regulation. Biochemistry80: 1-7.
- Zhao Q (2013) Allodynamicregulation of protein activity. Quantum Matter 2:144-152
- Taylor SS, Buechler JA, Yonemoto W (1990) cAMP-dependent protein kinase: framework for a diverse family of regulatory enzymes. Annu Rev Biochem 59: 971-1005.
- Fersht A (1999) Structure and mechanism in protein science: a guide to enzyme catalysis and protein folding. Macmillan learning.
- Frauenfelder H, Sligar SG, Wolynes PG (1991) The energy landscapes and motions of proteins. Science 254: 1598-1603.
- Shukla AK, Xiao K, Lefkowitz RJ (2011) Emerging paradigms of β-arrestin-dependent seven transmembrane receptor signaling. Trends BiochemSci 36: 457-469.
- Zimanyi CM, Chen PYT, Kang G, Funk MA,Drennan CL (2016) Molecular basis for allosteric specificity regulation in class Iaribonucleotidereductase from Escherichia coli. eLife 5: e07141.
- Zhao Q (2013) A molecular and biophysical model of the biosignal. Quantum Matter 2: 9-16.








