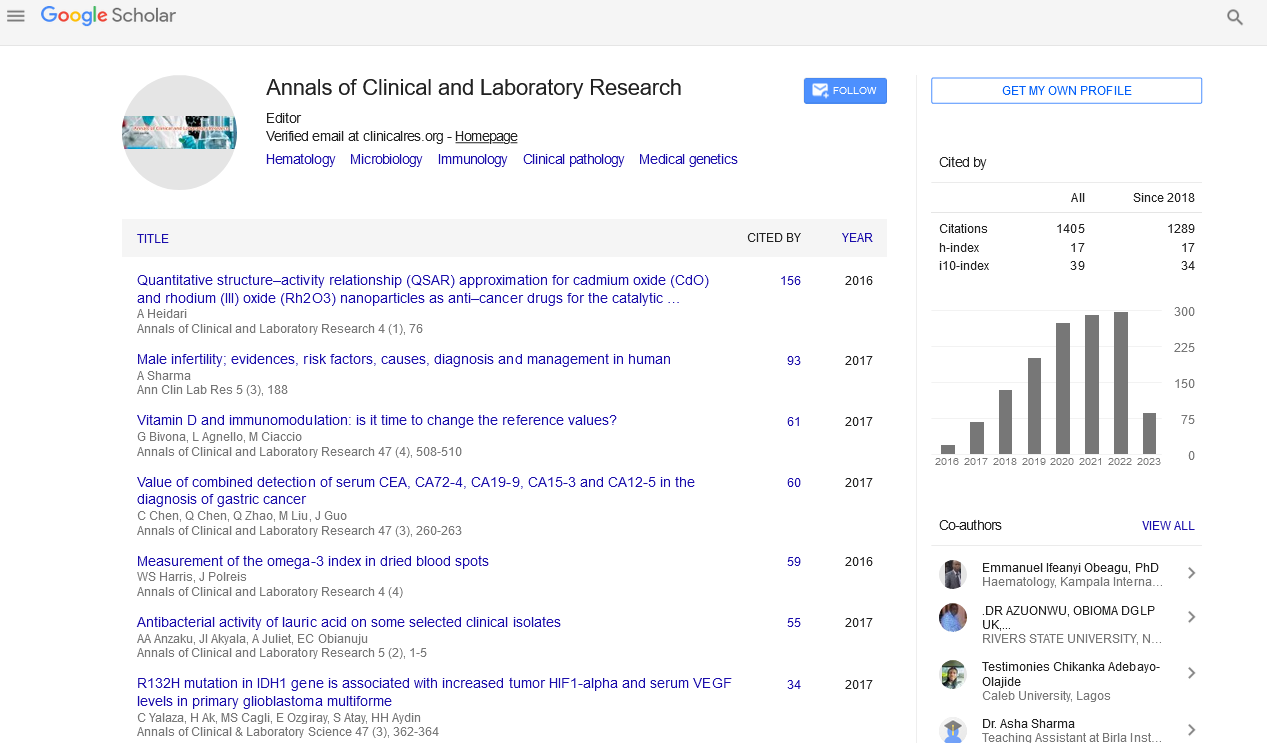
Perspective - (2023) Volume 11, Issue 1
Clinical Significance of Innate immunity on SARS-CoV-2
Kristin James*
Division of Epidemiology and Biostatistics, Faculty of Medicine and Health Sciences, Stellenbosch University, Cape Town, South Africa
*Correspondence:
Kristin James, Division of Epidemiology and Biostatistics, Faculty of Medicine and Health Sciences, Stellenbosch University,
South Africa,
Email:
Received: 04-Jan-2023, Manuscript No. IPACLR-23-13435;
Editor assigned: 06-Jan-2023, Pre QC No. IPACLR-23-13435;
Reviewed: 20-Jan-2023, QC No. IPACLR-23-13435;
Revised: 23-Jan-2023, Manuscript No. IPACLR-23-13435;
Published:
30-Jan-2023, DOI: 10.36648/2386- 5180.23.11.452
Abstract
People around the world have been impacted by the coronavirus disease 2019 (COVID-19) pandemic, which is brought on by the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). This has sparked an unprecedented effort from the scientific community to comprehend the biological basis of COVID19 pathophysiology. An overview of the current understanding of the innate and adaptive immune responses induced by SARS-CoV-2 infection as well as the immunological pathways that most likely influence the severity and mortality of the disease.
Keywords
ARS-CoV-2, Immunological pathways, Mortality, Biological basis.
Introduction
To immunity to viruses, innate immune sensing acts as the initial
line of antiviral defence. Very little about the precise innate
immune response to SARS-CoV-2 has been known. Given the
common sequence similarity across CoVs and the preserved
mechanisms of innate immune signalling, SARS-CoV-2 virus-host
interactions are anticipated to replicate many of those involving
other CoVs. When it comes to RNA viruses like SARS-CoV-2, these
pathways are started by viral single-stranded RNA (ssRNA) and
double-stranded RNA (dsRNA) engaging Pattern-Recognition
Receptors (PRRs) via cytosolic RIG-I like receptors (RLRs) and
extracellular and endosomal Toll-like receptors (TLRs).Following
PRR activation, cytokine production is triggered by downstream
signalling pathways. Type I/III Interferons (IFNs) are thought to
be the most crucial for antiviral defence among these, although
other cytokines are also generated, including proinflammatory
tumour necrosis factor alpha (TNF-alpha), Interleukin-1 (IL-1), IL-
6, and IL-18. They activate antiviral systems in target cells and
strengthen the immune response when combined. IFN-I can
successfully reduce CoV infection if it is detected early and is
targeted appropriately [1].
Coronaviruses have developed a number of methods to block
IFN-I production and signalling because these cytokines are a
significant barrier to viral infection. IFN secretion is suppressed
by SARS-CoV-1 both in vivo and in vitro, according to several
studies. The absence of significant type I/III IFN signals from infected cell lines, native bronchial cells, and a ferret model
suggests that SARS-CoV-2 likely has a similar impact. In contrast
to mild or moderate instances, individuals with severe COVID-19
have noticeably diminished IFN-I signatures. CoVs have a number
of evasion strategies, as is frequently the case. Viral elements
interfere with each stage of the process from PRR detection and
cytokine release to IFN signal transduction [2].
After activation, RLRs and TLRs trigger signalling cascades that
phosphorylate transcription factors including NF- κB and the IRF
family, which in turn triggers the production of proinflammatory
cytokines and IFN. The particular roles of the SARS-CoV-2 proteins
have not been defined experimentally; however proteomic
research has shown connections between viral proteins and PRR
signalling cascades. Through its interaction with Tom70, SARSCoV-
2 ORF9b indirectly interacts with the signalling adaptor
MAVS, supporting earlier studies that SARS-CoV-1 ORF9b inhibits
MAVS signalling [3]. Additionally, the SARS-CoV-2 NSP13 and
NSP15 interact with the signalling intermediate TBK1 and the
activator of TBK1 and IRF3 RNF41, respectively.
Proinflammatory and antiviral responses are out of balance:
When considered collectively, the variety of techniques used
by pathogenic CoVs to circumvent immune detection, notably
the IFN-I route, point to the crucial role that IFN-I response
dysregulation plays in COVID-19 pathogenicity. According to
animal models of both SARS-CoV-1 and MERS-CoV infection, the
severity of the illness is correlated with the inability to elicit an early IFN-I response. Perhaps more significantly, these models
show that timing is crucial since IFN is protective early in the
course of the disease but turns pathogenic later.
Prior Knowledge from Murine Coronaviruses, SARS-CoV-1, and
MERS-CoV Studies of myeloid cell dysfunction in SARS-CoV-1
and MERS-CoV can offer a crucial road map for understanding
COVID-19 pathogenesis even if information on COVID-19 patients
is still quickly emerging. Infection with SARS-CoV-1 causes an
abnormal AM phenotype in animal models, which inhibits DC
trafficking and T cell activation [4]. Furthermore, YM1+ FIZZ1+
alternative macrophages can heighten airway hypersensitivity,
aggravating fibrosis linked to SARS. Furthermore, as previously
mentioned, studies on the murine SARS-CoV-1 strain have shown
that inflammatory monocytes-macrophages and delayed IFN-I
signalling increase lung cytokine and chemokine levels, vascular
leakage, and impaired antigen-specific T cell responses, all of
which lead to lethal disease.
NK Cell Function is Affected by SARS-CoV-2 Infection ex vivo NK
cells from COVID-19 patients' peripheral blood had decreased
levels of CD107a, Ksp37, granzyme B, and granulysin intracellular
expression, which suggests poor cytotoxicity as well as impaired
synthesis of chemokines, IFN, and TNF. The deregulation of NK
cells may be caused by a number of routes [5]. Lung NK cells do
not exhibit the entrance receptor for SARS-CoV-2, ACE2, and are
therefore unlikely to be directly infected by SARS-CoV-2, while
influenza virus infects NK cells and causes apoptosis.
Conclusion
The scientific community has responded to the urgent need
for fundamental science and clinical research with exceptional
productivity due to the fast spread of SARS-CoV-2 and the
unusual character of COVID-19. The immunology of SARS-CoV-2 infections has been the subject of a sizable development of
scientific knowledge in recent months. Studies on the SARSCoV-
1 and MERS-CoV coronavirus epidemics in the past have laid
the groundwork for current knowledge. The immunopathologies
reported in SARS-CoV-1 and MERS-CoV infections, notably CRS,
are certainly present in severe COVID-19 patients. In addition to
reflecting our current knowledge that SARS-CoV-2 has a longer
incubation period and higher rate of transmission than other
coronaviruses, the emerging epidemiological observation that
significant proportions of people are asymptomatic despite
infection speaks to significant differences in the host immune
response. Therefore, it is essential that immune responses to
SARS-CoV-2 and the mechanisms of disease generated by hyper
inflammation are further understood to better define COVID-19
treatment options.
References
- Lowery SA, Sariol A, Perlman S (2021) Innate Immune and Inflammatory responses to SARS-CoV-2: Implications for COVID-19. Cell Host Microbe 14:1052-1062.
Indexed at, Google Scholar, Cross Ref
- Asselta R, Paraboschi EM, Mantovani A, Duga S (2020) ACE2 and TMPRSS2 Variants and Expression as Candidates to Sex and Country Differences in Covid-19 Severity in Italy. Aging (albanyNY) 6: 10087.
Indexed at, Google Scholar, Cross Ref
- Bacher P, Heinrich F, Stervbo U, Nienen M, Vahldieck M, et al. (2016) Regulatory T Cell Specificity Directs Tolerance versus Allergy against Aeroantigens in Humans. Cell 167:1067–1078.
Indexed at, Google Scholar, Cross Ref
- Brooks AG, Posch PE, Scorzelli CJ, Borrego F, Coligan JE (1997) NKG2A Complexed with CD94 Defines a Novel Inhibitory Natural Killer Cell Receptor. J Exp Med 185:795–800.
Indexed at, Google Scholar, Cross Ref
- Cao WC, Liu W, Zhang PH, Zhang F, Richardus JH (2007) Disappearance of Antibodies to SARS-Associated Coronavirus After Recovery. N Engl J Med 357:1162–1163.
Indexed at, Google Scholar, Cross Ref
Citation: James K (2023) Clinical Significance of Innate Immunity on SARS-CoV-2. Ann Clin Lab Res. Vol.11 No.1:452