Introduction
Listeria monocytogenes is a gram-positive bacterial pathogen and is the causative agent of the food-borne illness listeriosis. L. monocytogenes is responsible for nearly 28% of reported food-related deaths each year in the United States [1]. This bacterium is able to proliferate in a wide range of environments, making it a dangerous source of food contamination [2-5]. The implementation of a “zero-tolerance policy” for the presence of L. monocytogenes in ready-to-eat (RTE) food products in 1989 led to a drastic reduction in the incidence of L. monocytogenes contaminated RTE-products. However, there are still many reported cases of L. monocytogenes contamination and listeriosis annually [6], indicating that it is essential to understand the pathogenesis associated with this microbe.
Resistance to bile is considered a major virulence determinant for enterics [7]. The establishment of listeriosis is dependent upon the ability of L. monocytogenes to survive the acidic and bile environments encountered in the gastrointestinal system and to invade and replicate within the epithelial cells lining the intestinal tract [8]. Several mechanisms have been identified that allow for the resistance of L. monocytogenes to bile, including the bilE operon that excludes bile from the cytoplasm and bsh, which encodes a bile salt hydrolase involved in converting bile to a less toxic form [9-11]. In addition, the virulence regulator prfA regulates expression of these bile tolerance genes [9-11], indicating the necessity for bile resistance in the pathogenesis of L. monocytogenes. Bile is mainly composed of bile salts, cholesterol, and phospholipids. The detergent activity of bile is primarily attributed to the bile salt component [7]. L. monocytogenes is highly resistant to bile salts [9, 12]. However, it has not been analyzed whether this capability is directly related to pathogenicity. To determine if bile resistance could be related to the virulence capability of L. monocytogenes, we examined the effect that bile salts have on the survival and maintenance of the membrane integrity of the virulent strain EGD-e and the naturally isolated avirulent strain HCC23 under aerobic and anaerobic conditions. EGD-e is a serovar 1/2a strain that was initially described in 1926 following an outbreak of illness among laboratory animals [13]. HCC23 is a serovar 4a strain that was initially isolated from the brain of a healthy channel catfish and was subsequently found to be avirulent through mouse infection studies [14]. We found that bile salts affected the membranes of both EGD-e and HCC23, but this effect was much more severe in the avirulent strain. These results suggest that the pathogenic potential of L. monocytogenes could be directly related to bile tolerance, but bile tolerance should not be used as a sole indicator for virulence capability. We present these data and suggest a model for how bile may act as a bacteriostatic agent against L. monocytogenes.
Methods
Bacterial strains and anaerobic growth conditions. The L. monocytogenes strains EGD-e and HCC23 were grown in brain heart infusion (BHI) media at 37°C. All experimental methods were performed with cultures grown either aerobically or anaerobically. Anaerobic conditions were achieved by placing Wheaton serum bottles containing 1 ml of bile-infused BHI media in a vinyl anaerobic chamber for 48 hr (Type B, Coy Laboratory Products, Inc.), after which bottles were capped with aluminum seals. Syringes were used to inoculate cultures and remove samples.
Anaerobic conditions for BHI plates were achieved by incubating the plates in a BBL Gas Pak System. Anaerotest strips were used to verify anaerobic conditions. Growth analysis in the presence of bile salts. Fresh overnight cultures of EGD-e or HCC23 were diluted 1:100 in 2 ml of BHI medium containing 0%, 10%, or 20% oxgall, sodium glycodeoxycholate (GDCA), or sodium taurodeoxycholate hydrate (TDCA) and were grown at 37°C in a shaking incubator under either aerobic or anaerobic conditions. Bile salts were purchased from Sigma Aldrich. For each time point, 2 μl of culture were used for OD600 measurements with a Nanodrop ND-1000. Pathlengths for the Nanodrop readings were adjusted in accordance with the manufacturer. Three independent experiments were averaged for each bile salt under both anaerobic and aerobic conditions. Additionally, cultures grown in 0%, 10%, or 20% oxgall, GDCA, or TDCA for 8 hr at 37°C were plated onto BHI agar and incubated overnight at 37°C. Analyses of growth on agar plates were performed for three, independent experiments under both aerobic and anaerobic conditions. Cultures treated with bile under anaerobic conditions were plated and incubated under anaerobic conditions.
Scanning electron microscopy. Fresh overnight cultures of EGD-e or HCC23 were diluted 1:100 in 2 ml of BHI media infused with either 0% or 20% oxgall. After a 6 hr shaking incubation at 37°C, cells were centrifuged at 8,000 x g for 3 min at room temperature (Eppendorf Centrifuge 5415R). The resulting bacterial cell pellets were fixed in 2.5% (v/v) glutaraldehyde in 0.1 M cacoldylate buffer, washed in 0.1 M cacoldylate, post-fixed in 1% (v/v) osmium tetraoxide in 0.1 M cacoldylate buffer, rewashed in distilled water, dehydrated in an ethanol series, and dried in an hexamethyldisilazane (HMDS) series. Samples were sputter coated with goldpalladium (Polaron SEM coating system) prior to observation with a field emission scanning electron microscope (JEOL JSM-6500F).
Samples were prepared from three independent experiments of EGD-e or HCC23 treated with either 0% or 20% oxgall under both aerobic and anaerobic conditions. The length and width of 20 individual cells from each independent experiment were collected for analysis using the JEOL-PC-SEM 6500 software provided with the microscope. A mean average was calculated for cell length and cell width for data collected from the control cells (0% oxgall) and treated cells (20% oxgall). The mean average from the control cells was compared to the mean average from the bile treated EGD-e or HCC23 cells using a student t-test. A p-value < 0.05 indicated that the parameter measured (cell length or cell width) had significantly changed following treatment with oxgall.
Transmission electron microscopy. Sample preparations for TEM observations were performed as indicated above for the SEM with the following exceptions. After the dehydration step, the cells were treated in a stepwise resin/ethanol series and embedded into resin at 68 – 70°C overnight. The cells were then sectioned using an ultramicrotome (Reichert- Jung Ultracut E) and viewed under a transmission electron microscope (JEOL JEM-100CXII). The width of the cell wall, cell membrane, and cell envelope for 20 individual cells from each independent experiment was collected for analysis. Microscopic analyses were performed on three independent experiments for cells grown under aerobic or anaerobic conditions with either 0% or 20% oxgall.
A mean average was calculated for the cell wall, cell membrane, and cell envelope thickness for control cells (0% oxgall) and bile-treated cells (20% oxgall). The mean average from the control was compared to the mean average from the treated EGD-e or HCC23 using a student t-test. A p5 value < 0.05 indicated that the parameter measured (cell membrane, wall, or envelop thickness) had significantly changed following treatment with oxgall.
Results and Discussion
Analysis of EGD-e and HCC23 growth in the presence of bile salts
L. monocytogenes must be able to resist the damaging effects of bile salts in order to establish infections. With the recent finding that L. monocytogenes is able to replicate extracellularly within the gallbladder [15, 16] where bile salt concentrations can be 15% or higher, it has been suggested that resistance to high concentrations of bile salts is essential for the pathogenesis of this pathogen [7]. Additionally, it was recently found that bile aids in the formation of biofilms of L. monocytogenes [17], suggesting that this feature may allow for the survival of L. monocytogenes in the high bile environment of the gallbladder. To determine if the pathogenic potential of L. monocytogenes could potentially be related to bile tolerance, we examined the ability of the avirulent strain HCC23 and the virulent strain EGD-e to grow in the presence of 0%, 10%, or 20% of oxgall, GDCA, or TDCA under both aerobic and anaerobic conditions.
Under aerobic conditions growth was affected for both EGD-e and HCC23 in the presence of oxgall, TDCA, and GDCA (Fig. 1A, data not shown for TDCA and GDCA). The growth of HCC23 in the presence of bile salts was much more impaired than that observed for EGD-e. Increasing the concentration of bile salts exaggerated the growth deficiency exhibited by HCC23 (Fig. 1A). However, the avirulent strain did appear to maintain minimal cell viability through spectrophometric analysis. To determine if HCC23 was still viable following exposure to bile salts, cells were plated after 8 hr of incubation in the presence of oxgall, TDCA, or GDCA. Our results indicated that approximately 5% of the HCC23 cells remained viable in the presence of 20% oxgall as compared to untreated controls (3.8 x 108 treated with 20% oxgall as compared to 6.7 x 109 untreated viable at 8 hr).
The growth of EGD-e in the presence of bile salts was also impaired as compared to the control EGD-e cells (Fig. 1A). However, in contrast to HCC23 bile-exposed cells, growth of EGD-e increased to the level of control cells following an extended lag period (Fig. 1A). Under normal growth conditions, EGD-e exhibited a 1 hr lag phase. In the presence of 10% oxgall, TDCA, or GDCA this phase was extended to 2 hr. In the presence of 20% bile salts, this growth period was extended to 6 hr (Fig. 1A). The effect of the bile salt on EGD-e and HCC23 was similar regardless of whether oxgall or conjugated GDCA or TDCA was used as the treatment (data not shown).
To determine if similar growth patterns were observed in environmental conditions that potentially mimic those found within the human digestive system, we analyzed the ability of EGD-e and HCC23 to grow in the presence of bile salts under anaerobic conditions (Fig. 1B). In general, the growth of both strains was less prolific under anaerobic conditions. Both strains exhibited a decrease in growth with an increase in the concentration of bile salt. EGD-e had a 2hr lag phase in BHI media under anaerobic growth. This was slightly longer that that observed for aerobic conditions. Treatment of EGD-e with 10% bile salts increased the lag phase to 4 hr. However, exposure to 20% bile salts had a greater inhibitory affect on the growth of EGD-e under anaerobic conditions (Fig. 1B). These results were further analyzed by plating cells following an 8-hr incubation in the presence of 20% bile salts on BHI agar under anaerobic conditions. Approximately 17% of EGD-e treated with 20% oxgall survived as compared to untreated controls cells (3.71 x 109 treated with 20% oxgall as compared to 2.24 x 1010 untreated viable at 8 hr).
HCC23 did not have a longer lag phase under anaerobic conditions as was observed with EGD-e. However, cell growth was never able to reach the same concentration as that observed under aerobic conditions. Treatment with 10% and 20% bile salts under anaerobic conditions produced similar growth patterns as observed under aerobic conditions for HCC23 (Fig. 1).
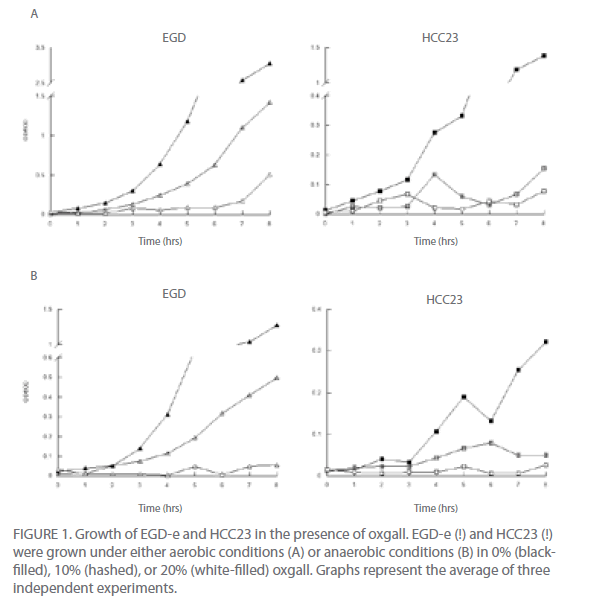
Figure 1: Growth of EGD-e and HCC23 in the presence of oxgall. EGD-e (!) and HCC23 (!) were grown under either aerobic conditions (A) or anaerobic conditions (B) in 0% (blackfilled), 10% (hashed), or 20% (white-filled) oxgall. Graphs represent the average of three independent experiments.
These results suggest that EGD-e may be better equipped to adapt to this stressful environment, repair DNA and membrane damage that might have been introduced by the bile salts, and resume replication to some extent.
Bile salts induce morphological changes in both EGD-e and HCC23
Utilizing scanning electron microscopy we were able to visually observe the different effects that bile salts have on the cell surface of these strains of L. monocytogenes. Since similar growth patterns were observed for oxgall, TDCA, and GDCA treated cells, only oxgall was used for morphological analyses. Additionally, since differences were seen in the ability of EGD-e to grow in the presence of 20% oxgall under different environmental conditions, morphological differences were analyzed in bile treated cells grown under aerobic and anaerobic conditions.
Changes in cell length and width were examined for three independent experiments for control and bile-treated EGD-e and HCC23 grown under aerobic and anaerobic conditions. In aerobic conditions nearly 78% of HCC23 treated with 20% oxgall exhibited visible surface deformities, indicating a loss of rigidity to the surface (Fig. 2A). These distortions were not present in the untreated HCC23, confirming that these alterations to the membrane were due to the bile salts. EGD-e showed less deterioration to the cell wall when exposed to oxgall; only 22% of the bile treated EGD-e cells exhibited damage to the cell surface under aerobic conditions (Fig. 2A). Additionally, these changes were not observed under control conditions. The length and width of control and bile-treated cells were measured to determine if oxgall altered the shape of L. monocytogenes under aerobic conditions. Oxgall did not significantly alter the cell length of either EGD-e or HCC23 (Table 1). However, the cell width of both strains was significantly altered in the presence of bile salts under aerobic conditions.
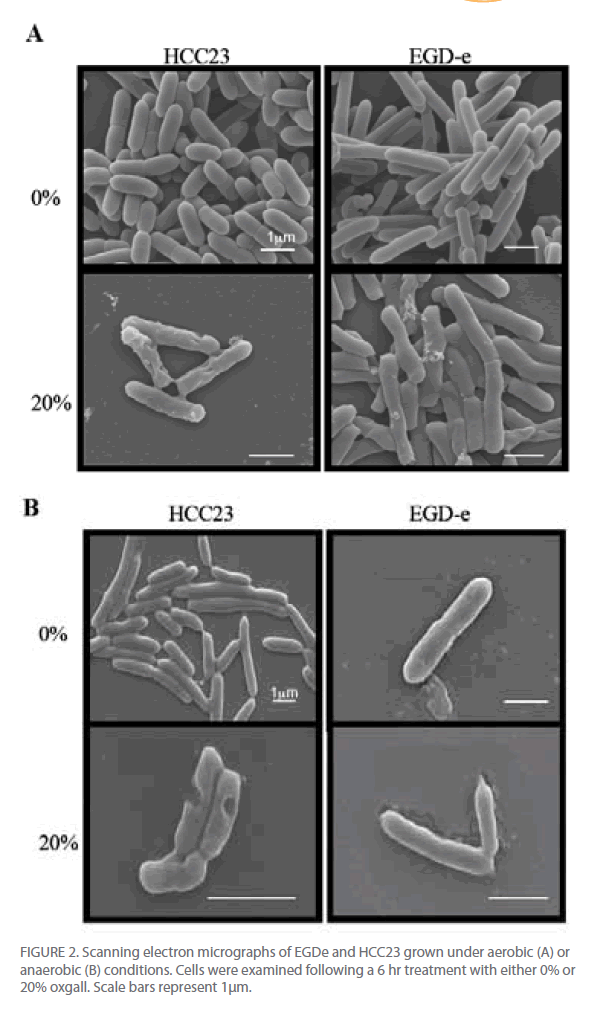
Figure 2: Scanning electron micrographs of EGDe and HCC23 grown under aerobic (A) or anaerobic (B) conditions. Cells were examined following a 6 hr treatment with either 0% or 20% oxgall. Scale bars represent 1μm.
The cell width of HCC23 decreased in the presence of 20% oxgall. Interestingly, the cell width of EGD-e significantly increased in the presence of oxgall (0.376 μm control v. 0.512 μm bile treated). This correlated with SEM micrographs that showed indentations along the surface of the cell wall in HCC23, while the rigidity and structure of the EGD-e cell membrane remained intact with little visible change (Fig. 2).
To determine if similar deformities occurred under anaerobic conditions, EGD-e and HCC23 treated with 20% oxgall under anaerobic conditions were observed by SEM. Similar to the observations under aerobic condition, bile salts induced significant amounts of damage to the surface of HCC23 (Fig. 2B). Under anaerobic conditions 73% of HCC23 cells exhibited cell surface deformities, while only 27% of EGD-e exhibited minimal damage. In anaerobic conditions growth in the presence of oxgall only significantly changed the cell morphology for HCC23 (Table 1). HCC23 exhibited a significant decrease in the cell length (1.518μm control v. 1.406μm bile treated) and cell width (0.503 μm control v. 0.429 μm bile treated (Table 1). It is possible that HCC23 becomes shorter and thinner as cytoplasmic material is lost through the compromised membrane. This has been demonstrated in Lactobacilli and Bifidobacteria, where it has been shown that bile salts dissipate the transmembrane electron potential [18]. This disrupts the membrane integrity and allows the leakage of protons, potassium ions, and other cellular components out of the cell [18]. The fact that EGD-e expanded in cell length could indicate a mechanism utilized to keep the membrane intact.
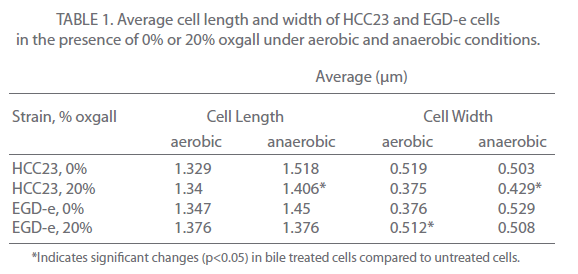
Table 1: Average cell length and width of HCC23 and EGD-e cells in the presence of 0% or 20% oxgall under aerobic and anaerobic conditions.
Bile salts alter the cell envelope of EGD-e and HCC23
To further investigate the effect that bile salts have on the cell envelope of EGD-e and HCC23, cells exposed to 20% oxgall under aerobic and anaerobic conditions were examined using a transmission electron microscope. Cells were analyzed for alterations in the thickness of the cell membrane, cell wall (peptidoglycan layer), and cell envelope (cell membrane and peptidoglycan layer). Comparing the thickness of the cell wall, membrane, and envelope between the control cells and biletreated cells of both strains revealed that bile affected the cell envelope of both strains under aerobic and anaerobic conditions (Fig. 3). Oxgall treated HCC23 and EGD-e grown under aerobic conditions showed a significant decrease in the average cell wall and cell envelope thickness when compared to their respective control (Table 2). EGD-e also had a significant decrease in the thickness of the cell membrane.
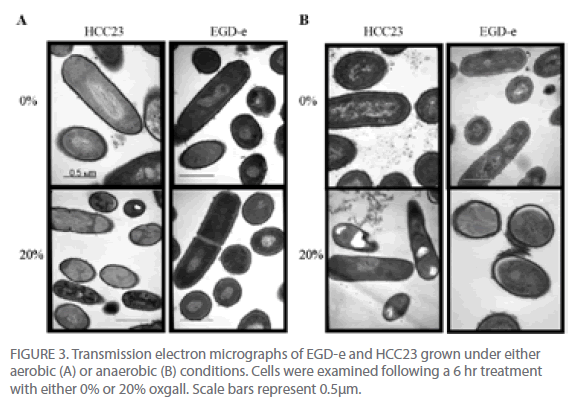
Figure 3: Transmission electron micrographs of EGD-e and HCC23 grown under either aerobic (A) or anaerobic (B) conditions. Cells were examined following a 6 hr treatment with either 0% or 20% oxgall. Scale bars represent 0.5μm.
Bile salts induced significant changes to the cell wall, membrane, and envelope of both strains under anaerobic conditions (Fig. 3B, Table 2). The average thickness of the HCC23 cell wall, membrane, and envelope significantly decreased when grown in the presence of bile under anaerobic conditions. While the cell wall and cell envelope of EGD-e decreased, the cell membrane thickness increased when grown in the presence of oxgall (3.5 nm control v. 5.74 nm bile-treated). This decrease in the thickness of the layers of the membrane was due, whether directly or indirectly, to the detergent effect of bile salts.
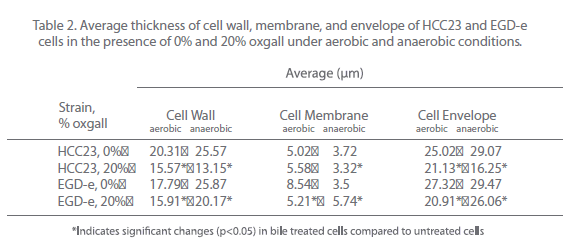
Table 2: Average thickness of cell wall, membrane, and envelope of HCC23 and EGD-e cells in the presence of 0% and 20% oxgall under aerobic and anaerobic conditions.
Even though the thickness of the cell envelope of EGD-e was significantly affected, the overall shape of the cell was not altered. This suggests that EGD-e has a mechanism for excluding the bile salts from the cell. Recently, a bile exclusion system (bilE) in L. monocytogenes was characterized for its ability to prohibit bile salts from entering the cell [11].
bilE expression was also shown to be regulated by the main virulence regulator prfA, which indicates that the exclusion of bile is related to virulence. Yet, even in this previous study, radiolabelled bile salts were still able to accumulate within the cell whether bilE was present or mutated, thus indicating that other mechanisms allow for the export of the bile once it has penetrated the membrane. Interestingly, HCC23 (NC_011660) also contains the bilE operon and shows 91% sequence similarity to the bilE gene in EGD-e (NC_003210). It also shows 97% similarity to the bsh gene of EGD-e (lmo2067) and 99% similarity to the proposed bile salt dehydrolase, btlB gene (lmo0754). HCC23 lacks pva (lmo0446), which is the only other gene identified for bile resistance in L. monocytogenes. This suggests that there may be other genes in EGD-e that are yet to be characterized for their involvement with tolerance and adaptation to bile. This area needs to be further explored in order to determine the factors that contribute to bile resistance.
Visible differences were also observed in the nucleoid and the cytoplasm of EGD-e and HCC23 treated with 20% oxgall (Fig. 3). Under aerobic conditions, 28% of HCC23 bile-treated cells appeared to have fragmented DNA (Fig. 3A). This affect was exaggerated under anaerobic conditions, where nearly 54% of the HCC23 bile-treated cells exhibited damage. Under both aerobic and anaerobic conditions, only 9% of EGD-e exhibited this same nucleoid damage. Cells were also seen to have areas where the cell membrane was dissociated from the cytoplasm. This was observed in 8% of HCC23 and only 2% of EGD-e under both aerobic and anaerobic growth conditions (Fig. 3). The areas where the membrane seemed to be dissociated from the cytoplasm correlated with the invaginations observed through the SEM. This could be due to the compromised membrane due to displacement of the phospholipids by the bile salts. However, it cannot be excluded that the morphological changes observed at the membrane could be due to autolytic enzymes activated through programmed cell death and are therefore not directly caused by the bile salts [19].
HCC23 accumulates bile salts within the cytoplasm
TEM micrograph examinations also indicated that HCC23 had patterns of intracellular darkening following 6 hr of exposure to oxgall. To determine if this cytoplasmic defect could be attributed to an influx of bile salts, we examined HCC23 following a 3-hr and 6-hr exposure to 0% or 20% oxgall in anaerobic conditions. TEM micrographs indicated that a visible darkening occurred within the cytoplasm of these cells. After 3 hr of bile exposure, approximately 13% of the treated HCC23 cells exhibited areas of partial darkening in the cytoplasm (Fig. 4). By 6 hr 20% of HCC23 exhibited this phenotype. HCC23 cells that were not exposed to oxgall did not have any areas of intracellular darkening within the cytoplasm (Fig. 4). Additionally, no areas of cytoplasmic darkening were observed within EGD-e in the presence or absence of bile salts (Fig. 3).
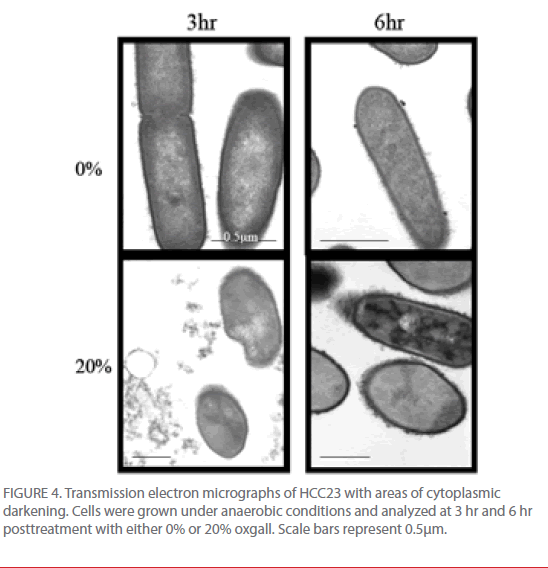
Figure 4: Transmission electron micrographs of HCC23 with areas of cytoplasmic darkening. Cells were grown under anaerobic conditions and analyzed at 3 hr and 6 hr posttreatment with either 0% or 20% oxgall. Scale bars represent 0.5μm.
The increase in intracellular darkening in the cytoplasm of HCC23 could indicate intracellular accumulation of bile salts. Because our SEM data indicated that the membrane of HCC23 was being significantly altered in the presence of bile salts it is no surprise that the accumulation of bile salts was specific to this strain. Bile salts that are able to breach the cell wall and accumulate within the cytoplasm could then exert DNA damage and arrest replication.
This idea is supported by the fact that more HCC23 cells had fragmented nucleoids compared to EGD-e. If DNA damage occurred the cell would become non-functional, which would explain the decrease in growth, especially in HCC23, in increasing concentrations of bile salts. However, it is also possible that the darkening is an artifact from processing the samples for TEM analysis. Regardless, this would still indicate that the HCC23 strain had a compromised membrane, as this intracellular darkening was not observed in EGD-e.
From these data and literature supporting the detergent properties of bile salts on lipids [20] , we propose a model for the potential effect of bile on the viability of L. monocytogenes (Figure 5). Bile salts act on the phospholipids and fatty acids of the membrane to disrupt the proton and potassium ion pumps, thus altering the transmembrane electron gradient and allowing for the influx of bile salts into the cell and efflux of cellular components out of the cell. Bile salts move into the cytoplasm and are targeted to the nucleoid. The bile salts then induce DNA damage, most likely through reactive oxygen species. If the damage is too profound for repair, the cell will cease to replicate. DNA damage induced by bile salts is well described in gramnegative bacteria [21-23] but remains in its infancy in gram-positive bacteria, though a recent study did find the induction of the nucleotide excision repair gene uvrA in L. monocytogenes cells in presence of bile salts [24]. The effect that bile has on the DNA of L. monocytogenes needs to be further analyzed.
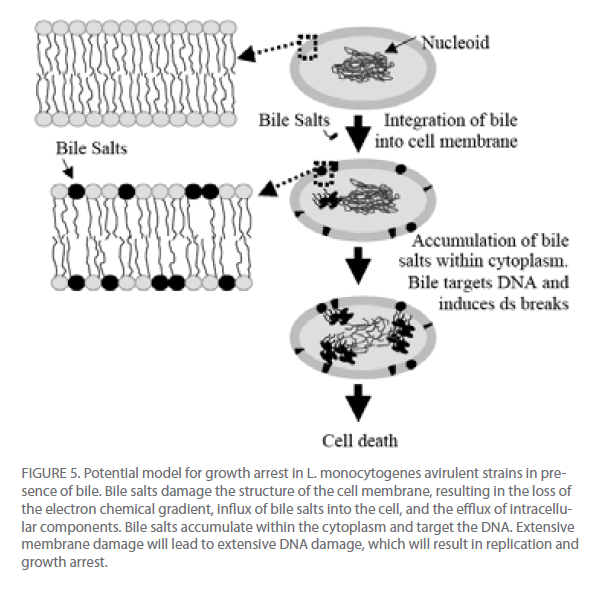
Figure 5: Potential model for growth arrest in L. monocytogenes avirulent strains in presence of bile. Bile salts damage the structure of the cell membrane, resulting in the loss of the electron chemical gradient, influx of bile salts into the cell, and the efflux of intracellular components. Bile salts accumulate within the cytoplasm and target the DNA. Extensive membrane damage will lead to extensive DNA damage, which will result in replication and growth arrest.
Acknowledgments
This work was supported by research funds provided to JRD by the Department of Biological Sciences and the Research Initiation Program at Mississippi State University. We would also like to thank Chris Holly and Cassandre Man-Bourdon for help with the EM analyses.
313
References
- Mead, P.S., Slutsker,L., Dietz, V., McCaig, L.F.,Bresee,J.S., Shapiro, C., Griffin,P.M. and Tauxe, R.V., Food-Related Illness and Death in the United States. Emerging Infectious Diseases, 1999. 5(5): p. 607-625.
- Cataldo, G., et al., Acid adaptation and survival of Listeria monocytogenes in Italianstylesoft cheeses. J ApplMicrobiol, 2007. 103(1): p. 185-93.
- Farber, J.M. and P.I. Peterkin, Listeria monocytogenes, a food-borne pathogen. MicrobiolRev, 1991. 55(3): p. 476-511.
- Gahan, C.G., B. O’Driscoll, and C. Hill, Acid adaptation of Listeria monocytogenescan enhance survival in acidic foods and during milk fermentation. Appl Environ Microbiol, 1996. 62(9): p. 3128-32.
- Graves, L.M. and B. Swaminathan, PulseNet standardized protocol for subtyping Listeria monocytogenes by macrorestriction and pulsed-field gel electrophoresis. Int J Food Microbiol, 2001. 65(1-2): p. 55-62.
- Crum, N.F., Update on Listeria monocytogenes infection. CurrGastroenterol Rep, 2002. 4(4): p. 287-96.
- Gunn, J.S., Mechanisms of bacterial resistance and response to bile. Microbes Infect,2000. 2(8): p. 907-13.
- Gahan, C.G. and C. Hill, Gastrointestinal phase of Listeria monocytogenes infection. J ApplMicrobiol, 2005. 98(6): p. 1345-53.
- Begley, M., et al., Contribution of three bile-associated loci, bsh, pva, and btlB, to gastrointestinal persistence and bile tolerance of Listeria monocytogenes. Infect Immun, 2005. 73(2): p. 894-904.
- Dussurget, O., et al., Listeria monocytogenes bile salt hydrolase is a PrfA-regulated virulence factor involved in the intestinal and hepatic phases of listeriosis. MolMicrobiol, 2002. 45(4): p. 1095-106.
- Sleator, R.D., et al., A PrfA-regulated bile exclusion system (BilE) is a novel virulence factor in Listeria monocytogenes. MolMicrobiol, 2005. 55(4): p. 1183-95.
- Begley, M., C.G. Gahan, and C. Hill, Bile stress response in Listeria monocytogenesLO28: adaptation, cross-protection, and identification of genetic loci involved in bile resistance. Appl Environ Microbiol, 2002. 68(12): p. 6005-12.
- Murray, E.G.D., R.A. Webb, and M.B.R. Swann, A disease of rabbit characterized by a large mononuclear leucocytosis, caused by a hitherto undescribed bacillus: Bacterium monocytogenes (n. sp.). J Pathol.Bacteriol., 1926. 29: p. 407-439.
- Erdenlig, S., A.J. Ainsworth, and F.W. Austin, Pathogenicity and production of virulence factors by Listeria monocytogenes isolates from channel catfish. J Food Prot, 2000.63(5): p. 613-9.
- Hardy, J., et al., Extracellular Replication of Listeria monocytogenes in the Murine Gall Bladder. Science, 2004. 303(5659): p. 851-853.
- Hardy, J., J.J. Margolis, and C.H. Contag, Induced Biliary Excretion of Listeria monocytogenes. Infect. Immun., 2006. 74(3): p. 1819-1827.
- Begley, M., C. Kerr, and C. Hill, Exposure to bile influences biofilm formation by Listeria monocytogenes. Gut Pathog, 2009. 1(1): p. 11.
- Kurdi, P., et al., Mechanism of Growth Inhibition by Free Bile Acids in Lactobacilli and Bifidobacteria. J. Bacteriol., 2006. 188(5): p. 1979-1986.
- Lewis, K., Programmed death in bacteria. MicrobiolMolBiol Rev, 2000. 64(3): p. 503-14.
- Garidel, P., et al., Membranolytic activity of bile salts: influence of biological membrane properties and composition. Molecules, 2007. 12(10): p. 2292-326.
- Bernstein, C., et al., Bile salt activation of stress response promoters in Escherichia coli. CurrMicrobiol, 1999. 39(2): p. 68-72.
- Kandell, R.L. and C. Bernstein, Bile salt/acid induction of DNA damage in bacterial and mammalian cells: implications for colon cancer. Nutr Cancer, 1991. 16(3-4): p. 227-38.
- Prieto, A.I., F. Ramos-Morales, and J. Casadesus, Bile-induced DNA damage in Salmonella enterica. Genetics, 2004. 168(4): p. 1787-94.
- Kim, S.H., et al., Role of uvrA in the growth and survival of Listeria monocytogenesunder UV radiation and acid and bile stress. J Food Prot, 2006. 69(12): p. 3031-6.












