Keywords
Pili, Klebsiella, conjugation, adhesion
Introduction
Adherence of bacteria to mammalian cell surfaces, mediated by bacterial pili or fimbriae, has long been recognized. Pilimediated attachment is correlated to the virulence of the organism. The attachment is followed by colonization of the tissue, eventually leading to intracellular penetration by the bacteria [1].
Expression of type 3 fimbriae has been described for many Gram-negative pathogens and is commonly detected in isolates of the genera Klebsiella, Enterobacter, Serratia, and Proteus. These fimbriae are characterized by their ability to mediate agglutination of tannic acid-treated human erythrocytes and this hemagglutination (HA) occurs in the presence or absence of D-mannose. Since hemagglutinin was originally characterized in Klebsiella strains, the fimbrial adhesin has been referred to as the mannose-resistant, Klebsiella-like (Mr/K) hemagglutinin [8]. Several studies have clearly demonstrated that type 3 fimbriae also mediates various adherence functions such as binding to epithelial cells and extracellular matrix proteins for instance, collagen V [8, 11, 12]. A putative regulatory gene (mrk E) located upstream of mrk A has been described previously in K. pneumoniae. These genes have been shown to reside in multiple genomic locations, including the chromosome, conjugative plasmids, and within a composite transposon. Transfer of a mrk-containing conjugative plasmid to strains of Salmonella entrica serovar typhimurium, K. pneumoniae, and E. aerogenes species has also been demonstrated [8].
Taken together, these data strongly support the spread of the mrk genes between Gram-negative pathogens by lateral gene transfer. In the present study, we focused on the probability of adhesion transfer as well as the probability of cotransfer of resistance markers from multiresistant K. pneumoniae isolates to a plasmidless strain.
Subjects and Methods
A total of 33 clinical K. pneumoniae isolates were recovered from 40 different clinical samples obtained from the Clinical Pathology Laboratory, Tanta University Hospitals. The organisms were identified and speciated based on colony morphology and biochemical reactions [7]. An Escherichia coli isolate (L99) was used as a plasmid molecular weight marker. This isolate was obtained from Department of Microbiology, Faculty of Pharmacy, Tanta University. E. coli NRRL (B-3707) strain which is a plasmid-free, lactose fermentor, chromosomally- mediated rifampicin-resistant (F-, Lac+, Rifr) was used as a recipient strain for mating in this study. This strain was obtained from the open culture collection of Agriculture Research Service (ARS) of Microbial Genomics and Bioprocessing Research Unit, Peoria, USA. The study was conducted in the Department of Microbiology, Faculty of Pharmacy, Tanta University from June 2010 to July 2011.
CaCO-2 cells (ATCC HTB-37) were kindly donated by the Tissue Culture Department of the holding company for biological products and vaccines (VACSERA), Cairo, Egypt. Dulbecco›s Modified Eagle›s Medium (DMEM) containing 1% nonessential amino acids was purchased from Media Department, VACSERA, Cairo, Egypt. Six-well tissue culture plates, cell scrapers, cover slips and tissue culture flasks (250 ml, 75 cm2, sterile, DNase, RNase free with filter cap) were purchased from Griener Bio-one, Germany.
Detection of virulent K. pneumoniae isolates
Preliminary screening of virulent K. pneumoniae isolates was done using Congo red binding assay as described by Berkhof and Vinal [1]. Briefly, K. pneumoniae isolates were streaked on MacConkey agar and incubated at 37oC for 24hrs. All the isolates were tested for their growth on trypton soy agar (Oxoid, UK) supplemented with 0.015 % bile salt and 0.03% Congo red. After 24 hrs of incubation, the cultures were left at room temperature for 48 hrs to facilitate the annotation of results. Virulent isolates were identified by their ability to bind to the Congo red dye and they appeared as red colonies while those that appeared white were considered negative.
Detection of adhesion as a virulence factor in virulent K. pneumoniae isolates
The adherence assay was carried out using the human cell line CaCO-2 cells as described by Di Martino et al [6]. Briefly, monolayers of differentiated CaCO-2 cells were prepared in 6-well tissue culture plates. The cells were seeded at 4x104 cells per cm2 in Dulbecco Modified Eagl’s Medium (DMEM) containing 20% fetal bovine serum (Lonza, Belgium), 10.000 U of penicillin per ml, 10 mg of streptomycin per ml, in an atmosphere of 10% CO2 at 37oC. Monolayers of CaCO-2 cells were used at semiconfluence; 3 days of culture. Cells were washed once with phosphate buffered saline (PBS, pH 7.2; 0.76% Na Cl, 0.07% Na2HPO4, 0.02% KH2PO4, Sigma USA). A suspension of 108 bacteria per ml of the cell line culture medium containing 0.5% (w/v) D-mannose was added to the tissue culture to inhibit type I pili and incubated for 3 hrs at 37oC. After three washes with PBS, the cells were fixed in methanol, stained with 20% Giemsa solution, and examined microscopically under oil immersion lens. An adhesion index was determined by examining 100 cells; it corresponded to the mean number of bacteria per cell. The results were expressed as means ± standard deviations. Each adhesion index represents the results of four separate experiments.
Susceptibility of the selected isolates to different antimicrobial agents
Susceptibility of CaCO-2 adherent K. pneumoniae isolates to different antimicrobials was performed using agar diffusion technique and the results were interpreted according to the Clinical and Laboratory Standard Institute guidelines, CLSI [4]. All antimicrobials used were purchased from Sigma, USA except for imipenem, norfloxacin, ciprofloxacin, enoxacin, levofloxacin, Moxifloxacin (Merk-Sharp, USA) and amikacin that were purchased from (Bristol-Meyer Squibb, USA).
Plasmid DNA analysis
Plasmids of CaCO-2 adhering K. pneumoniae isolates were prepared and analyzed by alkaline lysis method described by Birnboim and Doly [2].
Conjugation study
Selected isolates were subjected to conjugation study as described by Yan et al. [14].
Detection of virulence factors and/ or antimicrobial resistance markers in transconjugants
Transconjugants were cultured onto the selective media containing break points of each of the other antimicrobials and incubated overnight. They were tested also for adhesion as a virulence factor. Plasmid analysis of the transconjugants was carried out as described before.
Curing of plasmids
Transconjugants harboring more than one plasmid were subjected to curing study using ethidium bromide according to the method of Darfeuille-Michaud et al. [5].
Detection of pili encoded adhesion using scanning electron microscopy (SEM)
Being of high adhesion index, the donor (dK120) isolate, as well as its transconjugant (tK120), capable of adhering to CaCO-2 cells were viewed by SEM. The procedures were done in electron microscope unit, Faculty of Medicine, Tanta University, Egypt.
Characterization of the detected pili using transmission electron microscopy (TEM)
K. pneumoniae donor (dK120), the corresponding E. coli transconjugant (tK120), the recipient strain (B-3707), were grown at 37oC on Muller Hinton agar, harvested in phosphate buffered saline (pH 7.2) as described by Di Martino et al. [6]. The procedures were done in the electron microscope unit, Faculty of Medicine, Tanta University, Egypt.
Statistical analysis
The data were analyzed with SPSS version 15 statistical software package, 2006; Chicago, USA. Means were compared using the Student’s t-test for dependent samples. The number of replicas was 12 per each treatment.
Results
Detection of virulent bacterial isolates
Out of the 33 K. pneumoniae, 25 (75.8%) virulent isolates showed positive results.
Detection of adherent K. pneumoniae isolates
As shown in Table 1, 3 out of 25 (12 %) Congo red positive K. pneumoniae isolates adhered to the intestinal cell line, with high adhesion index values. Moreover, the three isolates showed diffuse adhesion pattern where bacteria scattered over the cell surface, and also there were some bacteria adhered strongly to the plastic surface of the tissue culture wells or glass cover slips as shown in Fig. 1. K. pneumoniae (K120) isolate adhered to approximately 80% of CaCO-2 cells as shown in Fig.1. Moreover, this isolate showed the highest adhesion index (6.45 bacteria per cell) as shown in Table 1 . In addition, this adhesion was observed even in the presence of 0.5% D-mannose.
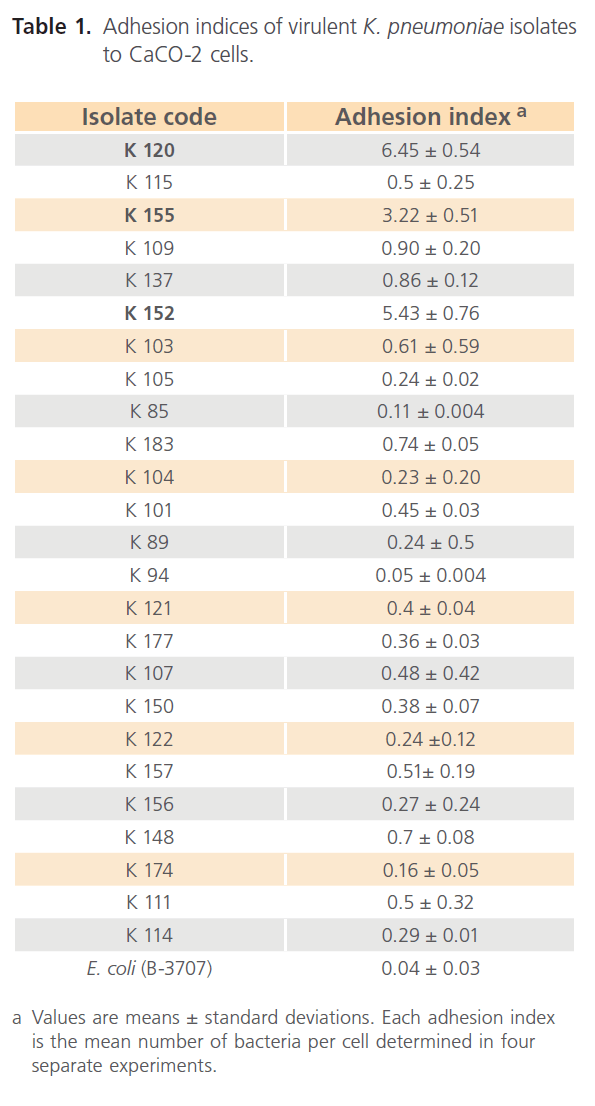
Table 1: Adhesion indices of virulent K. pneumoniae isolates to CaCO-2 cells.
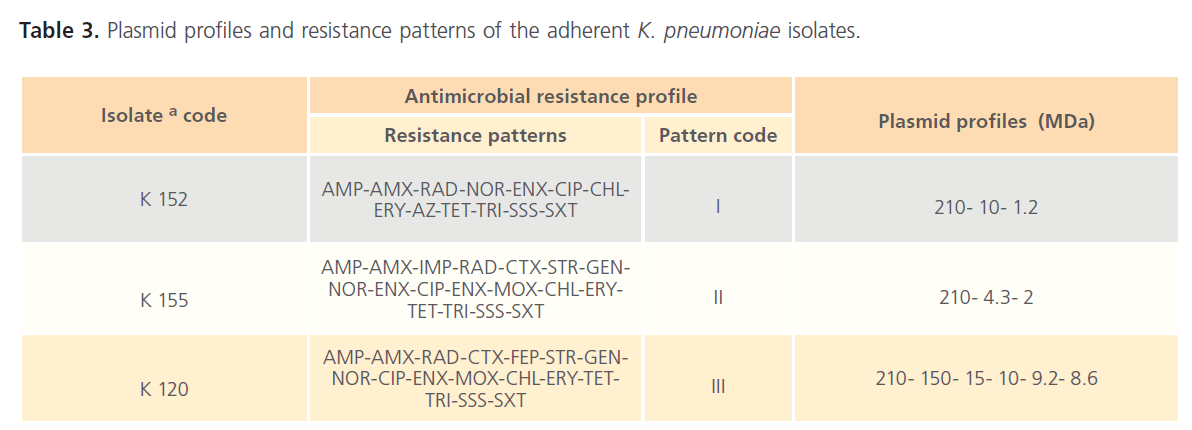
Table 3: Plasmid profiles and resistance patterns of the adherent K. pneumoniae isolates.

Fig 1: AdhesionLight micrographs of Giemsa–stained CaCO-2 cells showing (a) adherence of K. pneumoniae (K120) isolate, two rods of this isolate adhered to hairy-like structures on the surface of a CaCO-2 cell, (b) negative control of E. coli (B-3707). Magnification x100.
As shown in Fig. 2, electron micrographs of a negatively stained preparation of K120 isolate showed thin filamentous structures observed on its surface. These long, thin, and flexible fimbriae measure approximately 4 to 5 nm in diameter and 0.5 to 2 μm long as seen in (Fig 2 a). In contrast, E. coli (B-3707) recipient strain harbored no such pili (Fig. 2 c). Another isolate (K 103) of low adhesion index (0.61) was also examined. It showed a rigid and thick pilus of type I with a visible tip clearly seen in Fig. 2 b.
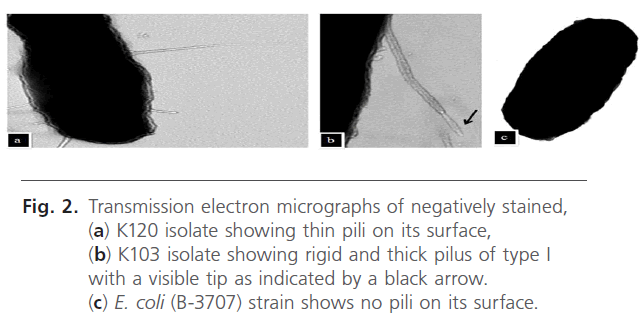
Fig 2: Transmission electron micrographs of negatively stained, (a) K120 isolate showing thin pili on its surface, (b) K103 isolate showing rigid and thick pilus of type I with a visible tip as indicated by a black arrow. (c) E. coli (B-3707) strain shows no pili on its surface.
Susceptibility of K. pneumoniae isolates to different antimicrobials:
The results shown in Table 2 describe the incidences of resistance of the three tested K. pneumoniae isolates to the various tested antimicrobials. The tested isolates were resistant to 50% of the tested antimicrobials including; AMP, AMX, RAD, TET, CHL, RIF, ERY, AZ, SSS, TRI, SXT, NOR, CIP and ENX. On the other hand, these isolates were susceptible to AMK and LEV.
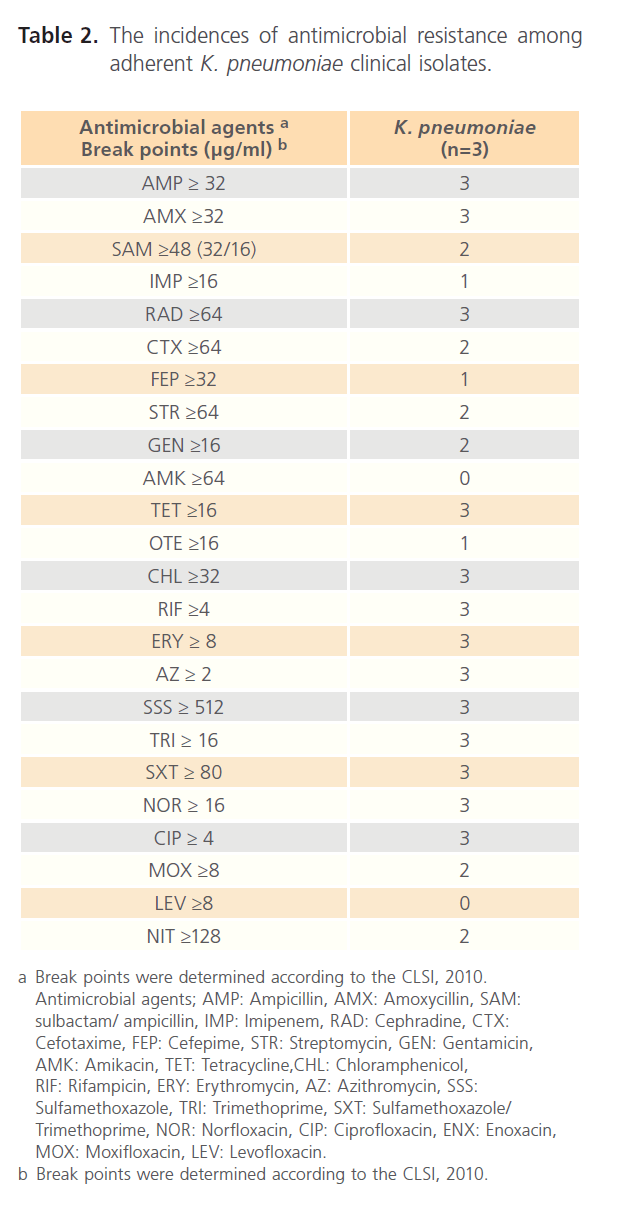
Table 2: The incidences of antimicrobial resistance among adherent K. pneumoniae clinical isolates.
Analysis of the plasmid profiles of adherent K. pneumoniae isolates
Plasmid profiles of the three K. pneumoniae isolates, capable of adhering to CaCO-2 cells, were presented in the electrophoregram shown in Fig. 3. As noted from Table 3, the isolates exhibited different resistance patterns and plasmid profiles. It was noted that a common plasmid with a large molecular weight (210 MDa) was present in the plasmid profile of the 3 tested isolates.
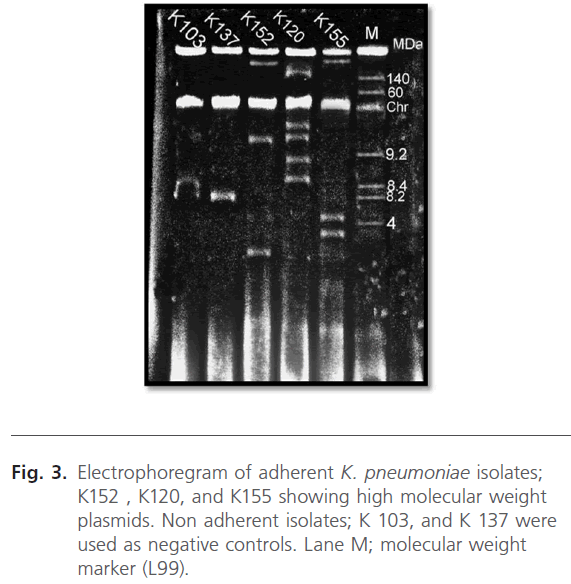
Fig 3: Electrophoregram of adherent K. pneumoniae isolates; K152 , K120, and K155 showing high molecular weight plasmids. Non adherent isolates; K 103, and K 137 were used as negative controls. Lane M; molecular weight marker (L99).
Transfer of adhesion encoding plasmids of K. pneumoniae isolates
Plasmid profiles of the donor isolates, their transconjugants and the selected cured derivative were presented in the electrophoregram shown in Fig. 4. Conjugation experiment revealed transfer of 210 MDa plasmid to the tested transconjugants as indicated in Fig. 4 and Table 4. Moreover, another 5 plasmids; 150, 15, 10, 9.2, and 8.6 MDa were cotransfered to tK120 strain as recorded in Table 4.
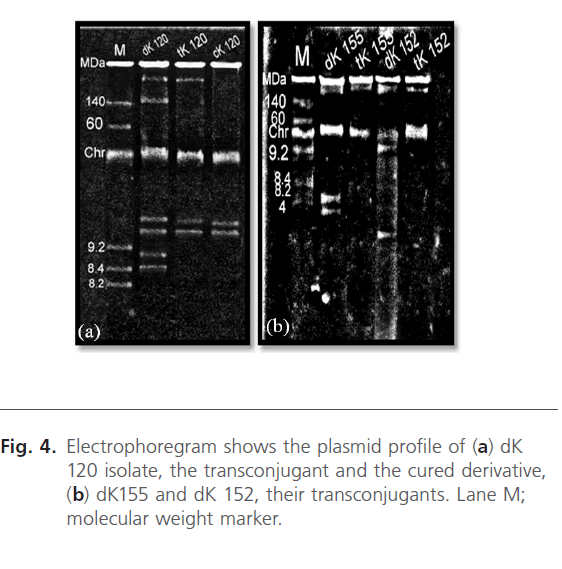
Fig 4: Electrophoregram shows the plasmid profile of (a) dK 120 isolate, the transconjugant and the cured derivative, (b) dK155 and dK 152, their transconjugants. Lane M; molecular weight marker.
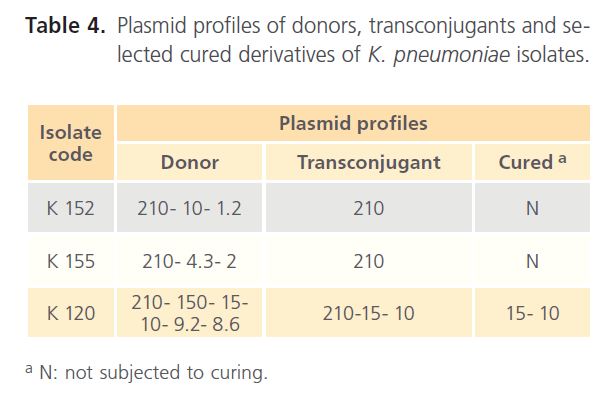
Table 4: Plasmid profiles of donors, transconjugants and selected cured derivatives of K. pneumoniae isolates.
Resistance markers transfer
As presented in Table 5, the resultant transconjugants acquired resistance to AMP, NOR, ENX, and CIP. In addition, curing experiment revealed that cK120 was still resistant to CHL and TET.
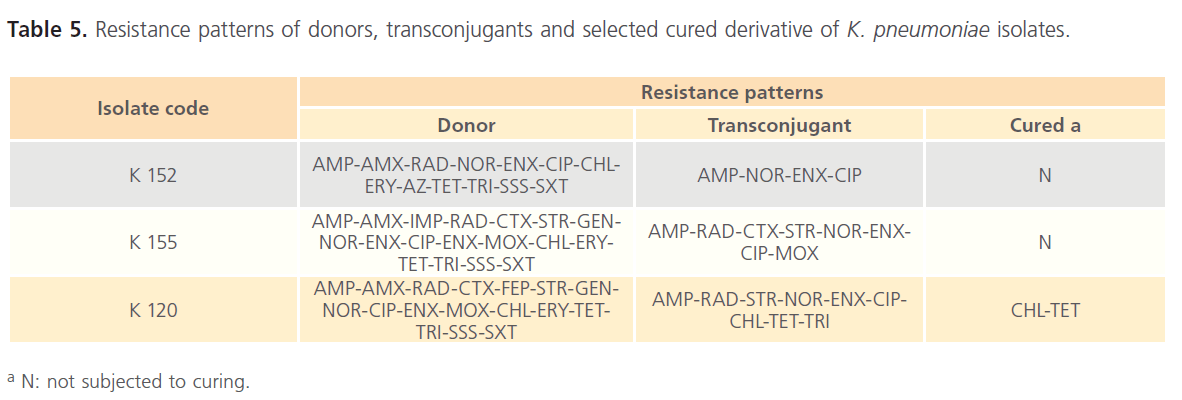
Table 5: Resistance patterns of donors, transconjugants and selected cured derivative of K. pneumoniae isolates.
Virulence factor transfer
Table 6 recorded that all transconjugants were capable of adhering to the CaCO-2 cells. The adhesion indices of the donors, their transconjugants and the selected cured derivative are shown in Table 7. All transconjugants showed high adhesion indices particularly tK120 that showed the highest value (8.69) and it was higher than its donor (6.45).
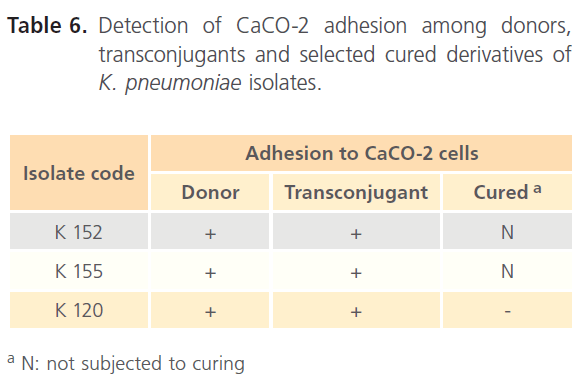
Table 6: Detection of CaCO-2 adhesion among donors, transconjugants and selected cured derivatives of K. pneumoniae isolates.
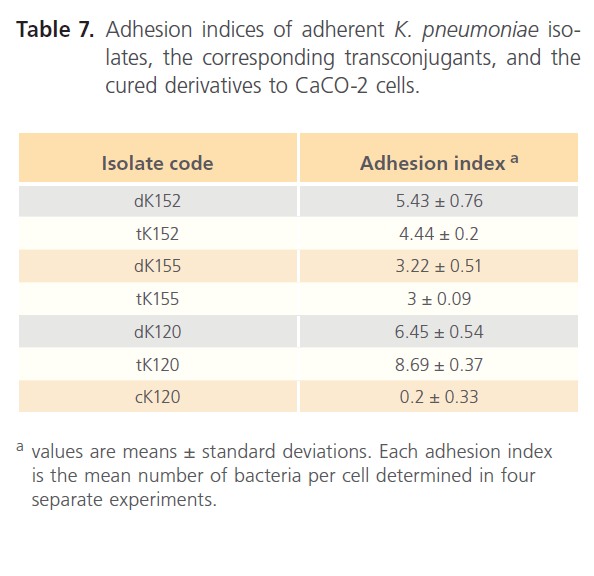
Table 7: Adhesion indices of adherent K. pneumoniae isolates, the corresponding transconjugants, and the cured derivatives to CaCO-2 cells.
Detection of adhesion of dK120 and tK120 to CaCO-2 monolayer by scanning electron microscope
It was found that dK 120 isolate, selected due to highest adhesion index, adhered to the surface of CaCO-2 via pili compared to the nonadherent E. coli B-3707 Fig. 5 a. This isolate can also adhere to the cytoskeletal fibers protruding from CaCO-2 cells. Moreover, this virulence factor was gained by the corresponding transconjugant tK 120 and pili were clearly seen in Fig. 5 b.
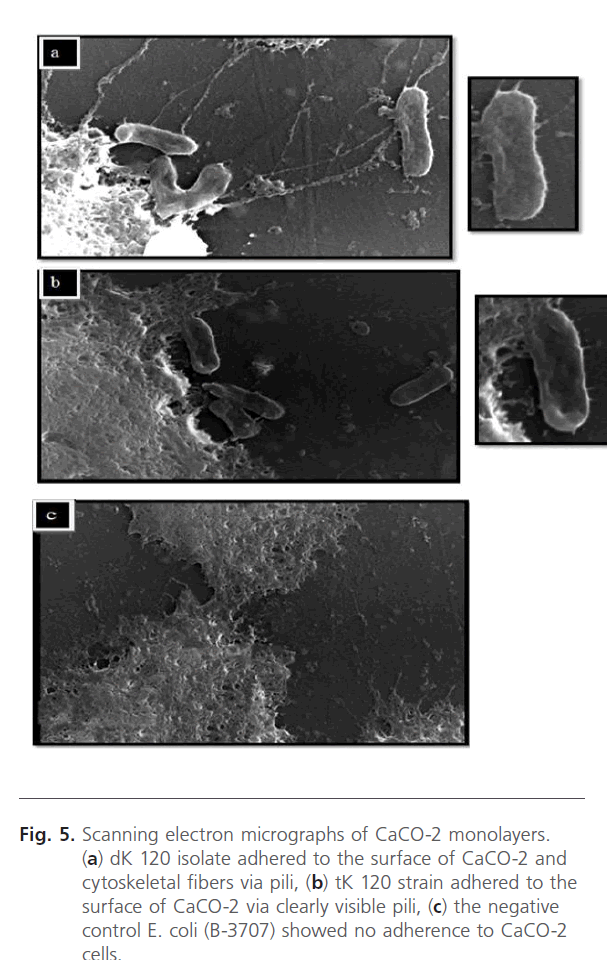
Figure 5: Scanning electron micrographs of CaCO-2 monolayers. (a) dK 120 isolate adhered to the surface of CaCO-2 and cytoskeletal fibers via pili, (b) tK 120 strain adhered to the surface of CaCO-2 via clearly visible pili, (c) the negative control E. coli (B-3707) showed no adherence to CaCO-2 cells.
Characterization of the pili of dK120 and tK120 using transmission electron microscope:
Electron micrographs of a negatively stained preparation of dK120 showed numerous filamentous structures on the bac bacterial surface that appeared to be more flexible and often form bundles along the whole surface of the bacteria. These long, thin, and flexible fimbriae measured approximately 4 to 5 nm in diameter and 0.5 to 2 μm long as observed in (Fig. 6 a). Similarly, the corresponding transconjugant tK120 harbored numerous longer fine pili protruding from the bacteria as shown in Fig. (6 b).
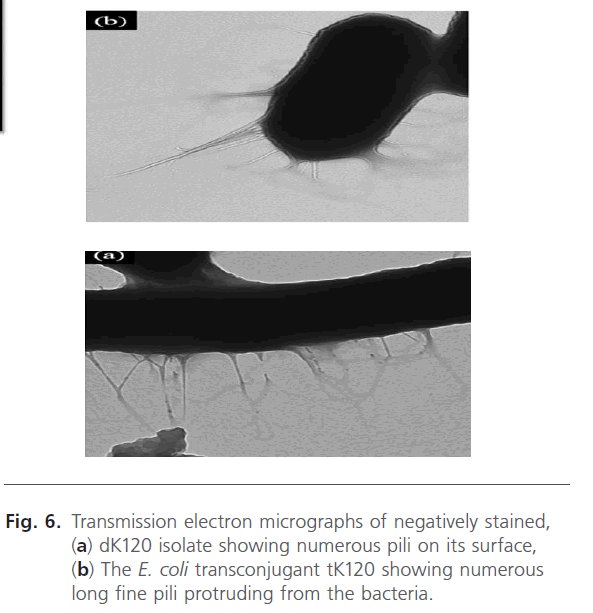
Figure 6: Transmission electron micrographs of negatively stained, (a) dK120 isolate showing numerous pili on its surface, (b) The E. coli transconjugant tK120 showing numerous long fine pili protruding from the bacteria.
Discussion
K. pneumoniae is an important Gram- negative opportunistic pathogen involved in hospital outbreaks of nosocomial infections including; acquired urinary tract infections, pneumonia, septicemia and infantile meningitis [5]. Virulence can be tested by Congo red test due to the presence of a strong correlation between expression of Congo red phenotype and virulence in avian E. coli. This might be associated with the presence of β-glucan in bacterial cell wall suggesting that Congo red binding can act as a virulence marker [1]. In our study, 25 out of 33 (75.8%) K. pneumoniae isolates showed positive Congo red. This was in agreement with the results of Shamlal et al., [10] who reported that in vitro pathogenicity testing of E. coli isolates revealed that 46 out of 97 (47.4%) of the isolates were positive for the Congo red binding.
Gastrointestinal acquisition and carriage of Klebsiella by the patient is an important intermediate step in the development of infections. This colonization process may be the result of the ability of Klebsiella to adhere to intestinal epithelial cells in the human gut [6]. To study the adhesiveness of Klebsiella, CaCO-2 cell line was used as a human intestinal model in the present study. It was found that 3 out of 25 (12%) virulent K. pneumoniae isolates showed positive adhesion to CaCO-2 cells. High adhesion indices were recorded particularly for K 120 isolate that showed the highest adhesion index (6.45 bacteria per cell) and this isolate was subjected for further study. Di Martino et al., [6] reported that 30 out of 78 (38.5%) K. pneumoniae isolates showed positive adhesion to CaCO-2 cells.
K. pneumoniae isolates involved in nosocomial infections are generally resistant to various antimicrobial agents such as penicillins, aminoglycosides, chloramphenicol, sulfonamides, tetracyclines, and broad spectrum cephalosporin [5, 13]. In the present study, more than 50% of the tested antimicrobial agents were inactive against the 3 tested K. pneumoniae isolates. These antimicrobials included; AMP, AMX, RAD, TET, CHL, ERY, AZ, SSS, TRI, SXT, NOR, CIP and ENX.
Darfeuille – Michaud et al. [5] and Schurtz et al. [9] reported the presence of 3 plasmids of size 7.6, 167, and 210 MDa in the test K. pneumoniae isolate. Conjugation and curing experiments revealed the transfer of 210 MDa plasmid conferring resistance to β-lactams, neomycin, streptomycin, sulfonamides and tetracyclines with a frequency approximately 10-6 to the recipient strain indicating that the resistance characteristics were all located on this conjugative plasmid. Moreover, the adhesion property of the donor isolate was also transferred. In addition, the adhesion index of the transconjugant was at least twofold higher than those of the wild-type K. pneumoniae isolate. There was an agreement between the results of Darfeuille – Michaud et al., [5] and the present study where a conjugative plasmid with a large molecular weight (210 MDa) was transferred from the three multiresistant K. pneumoniae isolates to their corresponding transconjugants following subjection to conjugation and curing experiments. This plasmid conferred resistance to AMP, RAD, NOR, ENX, and CIP. In addition, the transfer occurred with a very high frequency ranged between 0.45 x 10-3 to 1.6 x10-2 transconjugant per donor cell. Moreover, the adhesion property was transferred to the transconjugants particularly tK120 that showed adhesion index (8.69) higher than its donor (6.45) indicating that adhesion was plasmid- encoded. This finding was in agreement with Darfeuille – Michaud et al. [5] who reported that the difference in the level of expression of fimbrial protein between K. pneumoniae donor and E. coli transconjugant may result from the existence of genes involved in the regulatory process in K. pneumoniae. This would mean that these genes were not transferred or expressed in E. coli transconjugant, leading to high expression of that protein.s
The interaction of K. pneumoniae (dK120) isolate and E. coli transconjugant (tK120) with the CaCO-2 cells was analyzed by ultrastructure observations using scanning electron microscopy. It was noted that dK120 adhered to the surface of CaCO-2 via pili. These pili were clearly detected also in the transconjugant. Similarly, Di Martino et al., [6] reported that previously unidentified fimbriae, termed KPF-28, was involved in the adherence of pathogenic K. pneumoniae CF914-1 to the CaCO-2 cell line. On the other hand, Darfeuille – Michaud et al. [11] reported that adhesion of the tested K. pneumonia isolate to the intestinal cells was nonfimbrial.
Transmission electron microscope of a negatively stained preparation of both donor (dK120) and E. coli transconjugant (tK120), revealed elongated filamentous structures covering the surface. These structures appeared to be flexible, long, thin, and flexible fimbriae measuring approximately 3.5 to 4 nm in diameter and 1 to 2.5 μm long. Similar measurements were reported by Di Martino et al. [6]. It is formed by the polymerization of a 28-KDa major subunit. The native surface proteins extract of K. pneumoniae CF914-1 contained numerous fimbrial structures that aggregate with one another. Capetani et al. [3] reported that type 3 fimbriae produced by Enterobacteriaceae were morphologically similar and were characterized by their small diameter 2 to 4 nm and non channelled structure. Type 3 fimbrial adhesin gene (mrkD) of Klebsiella species is not conserved among all fimbriate strains. Previous report had indicated that fimbrial proteins of the P fimbria system were rapidly degraded by host proteases unless they were protected by chaperone proteins. Since the mrkD polypeptide can be incorporated into the P fimbrial filament, and because an expression vector carrying the P fimbrial chaperone gene papD has been constructed, mrkD expression was examined in transformants containing both mrkD and papD [9].
The present study correlated the virulence of K. pneumoniae with the presence of a 210 MDa plasmid encoding adhesive mannose-resistant pili which could adhere to the intestinal cell line. They were capable of strongly adhering to CaCO-2 cells, the cytoskeletal fibers, the glass cover slips, or even to the plastic surface of the tissue culture wells. Some resiss tance markers including; some β-lactams, flouroquinolones, and aminoglycosides were also encoded on a plasmid of size 210 MDa. It was evident that this plasmid was conjugative and hence could have a great impact on the dissemination of virulence factors as well as the resistance markers among bacteria facilitating the evolution of strains with enhanced virulence. Accordingly, it became important to search for adhesion and conjugation inhibitory compounds to be coadminstered to improve the antimicrobial therapy of K. pneumoniae infections.
124
References
- Berkhoff HA, Vinal AC. Congo red medium to distinguish between invasive and non-invasive E. coli pathogenic or poultry. Avian Dis. 1986; 30: 117-21.
- Birnboim HC, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979; 7: 1513-23.
- Capitani G, Eidam O, Glockshuber R, Grütter MG. Structural and functional insights into the assembly of type I pili from Escherichia coli. Microbes Infect. 2006; 8: 2284-90.
- CLSI/NCCLS. Performance standards for antimicrobial susceptibility testing: Twentieth informational supplement. NCCLS/CLSI/document M100-S20. Vol. 30(1): 41-44, 62-79. Franklin, R.C.; Matthew, A.W.; Karen, B.; Michael, N.; George, M.E.; Dwight, J.H.; David, W.H. et al. Clinical and Laboratory Standards Institute, 940 West Valley Road, Suite 1400, Wayne, Pennsylvania, USA. 2010.
- Darfeuille-Michaud A, Jallat C, Aubel D, Sirot D, Rich C, Sirot J, et al. R-plasmid-encoded adhesive factor in K. pneumoniae strains
- responsible for human nosocomial infections. Infect Immun. 1992; 60: 44-55.
- Di Martino P, Livrelli V, Sirot D, Joly B, Darfeuille-Michaud A. A new fimbrial antigen harbored by CAZ-5/SHV-4-producing Klebsiella pneumoniae strains involved in nosocomial infections. Infect Immun.1996; 64: 2266-73.
- Guo XL, Wang DC, Zhang YM, Wang XM, Zhang Y, Zuo Y, et al. Isolation, identification and 16S rDNA phylogenetic analysis of Klebsiella pneumonia from diarrhea specimens. Zhonghua Liu XingBing Xue Za Zhi 2008; 29: 1225-9.
- Ong CL, Ulett GC, Mabbett AN, Beatson SA, Webb RI, Monaghan W, et al. Identification of type 3 fimbriae in uropathogenic Escherichia coli reveals a role in biofilm formation. J Bacteriol. 2008; 190: 1054-63.
- Schurtz TA, Hornick DB, Korhonen TK, Clegg S. The Type 3 fimbrial adhesin gene (mrkD) of Klebsiella species is not conserved among all fimbriate strains. Infect Immun. 1994; 62: 4186-91.
- Shamlal R, Rajarathnam S, Sankaran K, Ramachandran V, Subrahmanyam YV, Nair GB, et al. Detection of virulent Shigella and enteroinvasive Escherichia coli by induction of the 43 kDa invasion plasmid antigen, ipaC. FEMS Immunol Med Microbiol. 1997; 17: 73-8.
- Tarkkanen AM, Allen BL, Westerlund B, Holthöfer H, Kuusela P, Risteli L, et al. Type V collagen as the target for type-3 fimbriae, enterobacterial adherence organelles. Mol Microbiol. 1990; 4: 1353-61.
- Tarkkanen AM, Allen BL, Williams PH, Kauppi M, Haahtela K, et al. Fimbriation, capsulation, and iron scavenging systems of Klebsiella strains associated with human urinary tract infection. Infect Immun. 1992; 60: 1187-92.
- Tasli N, Bahar IH. Molecular characterization of TEM- and SHV-derived extended-spectrum beta-lactamases in hospital-based Enterobacteriaceae in Turkey. Jpn J infect Dis. 2005; 58: 162-7.
- Yan JJ, Ko WC, Wu JJ. Identification of a plasmid encoding SHV-12, TEM-1, and a variant of IMP-2 metallo-ß-lactamase, IMP-8, from a clinical isolate of Klebsiella pneumoniae. Antimicrob Agents Chemother 2001; 45: 2368-71.


















