Pantopikou K and Papasotiriou I*
Research Genetic Cancer Centre SA (RGCC), Industrial Area of Florina, GR53100, Florina, Greece
- *Corresponding Author:
- Ioannis Papasotiriou
Head of Molecular Medicine at RGCC, Florina, Greece
Tel: +302385041950
Fax: +302385041931
Email: office@rgcc-genlab.com, papasotiriou.ioannis@rgcc-genlab.com
Received Date: April 29, 2017; Accepted Date: May 08, 2017; Published Date: May 15, 2017
Citation: Pantopikou K, Papasotiriou I. Detection and Identification of Bacterial Contamination in Blood Samples from Cancer Patients Arch Clin Microbiol. 2017, 8:3. doi: 10.4172/1989-8436.100042
Copyright: © 2017 Pantopikou K, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Keywords
Bacterial contamination; PCR; Gram+ve bacteria; Cancer patients; Blood samples
Introduction
Bacterial contamination of blood samples resulting from improper blood drawing technique or mishandling of samples by laboratory staff is a frequent problem in diagnostic laboratories. Factors potentially leading to bacterial growth include poor hand hygiene, lack of glove sterilization, insufficient skin disinfection or skin core removal of collected blood during phlebotomy. Gram-positive (Gram+) bacteria account for at least half of all documented microbiological infections in cancer patients [1]. Skin bacteria, such as Staphylococcus epidermidis (S. epidermidis), are the most frequent source of contamination. While these organisms do not typically grow at low temperatures (1-6°C), they survive and readily multiply when plated and stored at room temperature (20-24°C) [2]. Skin bacteria, such as Streptococcus pyogenes (S. pyrogenes), Staphylococcus aureus (S. aureus) and S. epidermidis, can contaminate blood samples. Moreover, bacteria frequently found in mammalian respiratory tracts, such as Streptococcus pneumoniae (S. pneumoniae), have also been reported to cause frequent infections in cancer patients [3]. Immunosuppression induced by the underlying cancer or its attendant therapy, such as neutropenia and breakdown of mucosal barriers during longterm vascular catheter placement or graft versus host disease, can synergize to render cancer patients particularly susceptible to Gram+ infections.
In this study, presence of the aforementioned skin and respiratory bacteria was investigated by polymerase chain reaction (PCR). To determine whether specimen contamination was caused by the physician during blood collection or by the laboratory environment, blood sampled directly from the blood collection tube was cultured in chocolate blood agar prior to DNA isolation. From the same collection tube, DNA was also isolated from plasma to assess staff handling. Bacterial presence or absence was determined by PCR amplification of speciesspecific genomic regions.
Material and Methods
Sample collection
Blood samples from patients diagnosed with cancer were collected in sterilized 50-ml Falcon conical tubes (Corning, NY, USA) containing 7 ml of 0.02 M ethylenediaminetetraacetic acid (EDTA; Duchefa Biochemie B.V., Haarlem, The Netherlands) as an anticoagulant. Total volume of collected blood was 20 ml. Samples were cultivated on a roller for 30 minutes and then sent to the laboratory for analysis. Time of sample transit to the laboratory did not exceed 72 hours. Samples were received both from USA and Europe.
Examination for bacterial contamination
For detection of contamination, 50 μl was collected from all patient samples and placed on petri dishes containing blood agar base (70133; Sigma-Aldrich, St. Louis, MO, USA), prepared according to the manufacturer’s instructions. Samples were incubated for 48 h at 37°C and then examined for contamination by determining the presence of bacterial colonies. Twenty-eight contaminated samples were randomly selected, and their DNA was isolated and analyzed by PCR of both colonies and serum.
DNA isolation
One bacterial colony was isolated from contaminated samples and resuspended in 20 mg/ml of lysozyme solution. Plasma from the same samples was isolated, centrifuged, and resuspended in lysozyme solution. All samples were subsequently incubated for 2 hours at 37°C. DNA extraction was carried out using a QIAamp DNA Mini Kit (51306; Qiagen, Hilden, Germany) following the protocol for Gram+ bacteria. Samples were then evaluated spectrophotometrically.
PCR amplification
Primers used for amplification of bacterial DNA were designed according to the literature (Table 1). Primers for S. aureus amplified an approximately 270-bp DNA fragment of the nuc A gene, which encodes nuclease A [4]. For S. pyogenes, primers amplified a 407-bp DNA fragment from the putative transcriptional regulator gene spy1258 [5]. For S. pneumoniae, primers amplified the capsulation locus cps; whereas, for S. epidermidis, the putative transcriptional regulator gene serp0107 was amplified [6]. One positive (commercial DNA for each bacterium) and two negative (no template, reference genomic DNA) controls were used for each reaction. PCR was performed in a total volume of 25 μl. The reaction mixture was composed of 0.5 units of Taq DNA polymerase (M8305; Promega, Madison, WI, USA), 1X PCR buffer, 0.4 mM primers, 2 mM MgCl2, 0.2 mM of each dNTP, and ddH2O. The cycling program consisted of one cycle at 95°C for 2 min; 35 cycles of 94°C for 15 s, 62.6°C for 20 s, and 72°C for 30 s; and one cycle at 72°C for 2 min. To validate the quality of PCR products, an extra PCR reaction was performed on all samples using universal primers for 16S rRNA. Primers used were based on the literature [7]. After the completion of all cycles, 2 μl of 6X DNA loading buffer (9156; Takara, Tokyo, Japan) was added to each tube and the resultant products were examined by 2% agarose gel electrophoresis in the presence of Midori Green (MG04; Nippon Genetics, Tokyo, Japan). A 100-bp ladder (MWD100; Nippon Genetics) was used to estimate the size of PCR products.
| Organisms |
Amplified gene |
primers |
length |
| Staphylococcus aureus |
nuc A |
5'-GCGATTGATGGTGATACGGTI-3'
5'GCCAAGCCTTGACGAACTAAAGC-3' |
270bp |
| Staphylococcus epidermidis |
serp0107 |
5’-TTGAGCTTGTCATTGGTTCG-3’
5’-TGTAGAGGTTGCACGTCGAG-3’ |
581bp |
| Streptococcus pyogenes |
Spy1258 |
5’-AAAGACCGCCTTAACCACCT-3’
5’-TGGCAAGGTAAACTTCTAAAGCA-3’ |
407bp |
| Streptococcus pneumoniae |
Cps2 |
5’GCAATGCCAGACAGTAACCTCTAT3’ 5’CCT GCC TGC AAGTCT TGA TT-3’ |
1061bp |
| Universal primers |
16S rRNA |
5'-AGGAGGTGATCCAACCGCA-3'
5'-AACTGGAGGAAGGTGGGGAT-3' |
351bp |
Table 1: Bacterial primers and amplified loci.
Results
Based on the results of electrophoresis, all samples positive for at least one bacterium yielded an appropriate band for 16S rRNA (351 bp), indicating good quality of extracted DNA. Among paired plasma-colony samples, 23 were contaminated with S. aureus, 12 with S. epidermidis, 12 with S. pneumoniae, and two with S. pyogenes (one for both colony and plasma, and one only for the colony) (Table 2). Some of these samples were contaminated with more than one bacterial strain (Figures 1 and 2). All expected bands were detected in positive controls, while bands were absent in negative controls. Faint bands appeared for Staphylococcus aureus, Streptococcus epidermidis and Streptococcus pneumoniae.
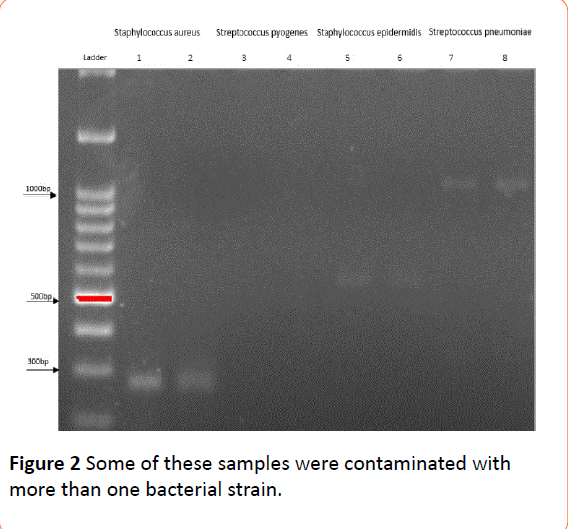
Figure 1: Some of these samples were contaminated with more than one bacterial strain.
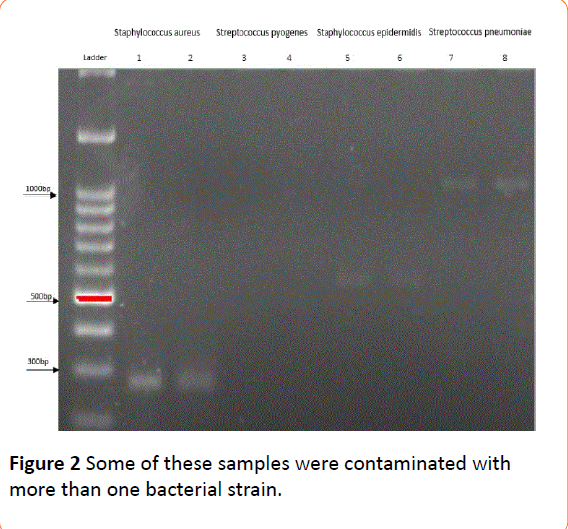
Figure 2: Some of these samples were contaminated with more than one bacterial strain.
| Species |
Plasma samples |
Samples from petri dish colonies |
Plasma/Colonies positive |
| Staphylococcus aureus |
82% |
82% |
100% |
| Streptococcus epidermidis |
43% |
43% |
100% |
| Staphylococcus pyogenes |
3.5% |
7% |
50% |
| Streptococcus pneumoniae |
43% |
43% |
100% |
Table 2: Percentage of samples contaminated with each bacterium.
Discussion
Blood culture is perhaps one of the most important laboratory procedures. However, contamination of blood samples with bacteria primarily derived from skin continues to be a frequent problem. Failure of the health worker to use strict aseptic technique when obtaining the blood specimen, or inappropriate sample handling by laboratory staff can lead to this problem. Alternatively, bacterial presence within samples may be independent of blood collection and handling, instead resulting from an underlying patient infection with the same bacteria. In this study, the presence of four Gram+ bacteria was investigated by PCR in blood samples collected from cancer patients.
Frequently found in the mammalian respiratory tract, S. pneumoniae is a part of the normal flora in small densities, however, when its population becomes too large, it essentially becomes pathogenic. Commonly found in pairs (diplococci), but also in isolation and short chains, S. pneumoniae are slightly pointed, cocci-shaped Gram+ bacteria characterized by a polysaccharide capsule that completely encapsulates cells.
S. pyogenes is a free-living organism, however, its ecological niche appears to be quite narrow: its only known biological host is human and, like many other streptococci, it lacks an environmental reservoir of known importance. Thus, direct human-to-human transmission, which mostly occurs via respiratory droplets or skin contact,is critical for its maintenance. Streptococcal organisms tend to reside extracellularly, but can also be found within mammalian host cells of the respiratory tract [8,9]. S. pyogenes is capable of causing a large variety of infections [10-18].
S. aureus is one of the most common hospital-acquired pathogens. As a normal inhabitant of skin and nasal mucous membranes of healthy humans that may only survive on dry skin, S. aureus is infectious to both animals and humans. It can be spread through contaminated surfaces, air and human contact. Approximately 30% of the normal healthy population is affected by S. aureus, as it asymptomatically colonizes the skin of human hosts. Although, host colonization can be benign, a puncture or break in the skin can prompt this bacterium to enter through the wound and cause infection. As S. aureus are spherical, Gram+, and immobile, they form grape-like clusters [19].
Usually originating from disease, S. epidermidis is a Gram+ staphylococcus that lives on human skin and mucosa. As one of the five most frequent causes of nosocomial infections, S. epidermidis is often compared with S. aureus; together, the two genus-sharing pathogens cause over one million serious hospital-acquired infections each year. S. epidermidis is the dominant staphylococcus species on skin, while S. aureus lives mostly on mucosal surfaces; however, when S. aureus is present, S. epidermidis decreases drastically. Multiple S. epidermidis strains from the nares of healthy adults show that many types of this particular organism exist within a single individual. Notably, the ability to produce slime allows S. epidermidis to adhere to biomaterials.
In this study, blood samples were received at the laboratory from USA and European countries; many samples were found to be contaminated. To define when the contamination may have occurred, a small volume of each blood sample was placed on blood agar dishes and incubated to investigate the presence of bacteria. Twenty-eight contaminated samples were randomly chosen and plasma from the corresponding blood-containing tube was isolated for investigation of bacterial presence. Results of PCR performed for both colonies and plasma indicated the most frequent contamination (82%) was caused by S. aureus, while 42%, of samples were contaminated with S. pneumoniae and S. epidermidis. Additionally, one sample was contaminated with S. pyogenes on the petri dish, but not in the corresponding plasma.
A systematic analysis of bacteremia studies among cancer patients found that S. aureus accounted for 1.3-12% of all cases [20]. Malignancy is also a risk factor for invasive disease secondary to S. pneumoniae, with patients exhibiting active leukemia, lymphoma or myeloma, or those having undergone stem cell transplantation, having the highest incidence [21]. While pneumonia is the most common source of S. pneumoniae bacteremia in cancer patients, other sources, including occult, have also been reported [22,23]. Although most invasive infections arising from S. pneumoniae in cancer patients are caused by serotypes found in major pneumococcal vaccines, being immune compromised is also a risk factor for invasive pneumococcal disease caused by non-vaccine covered serotypes [24].
While there are many ways to differentiate true bacteremia from blood culture contamination, such as microbe identification and corroboration of clinical and laboratory data, we cannot be certain of the source of contamination because the patient’s clinical condition and appropriateness of the laboratory environment where blood handling took place are unknown to us. Although, according to Weinstein’s study of 843 positive blood samples, certain organisms like S. pneumoniae and S. aureus almost always represent true bacteremia in a significant proportion of cases, in contrast to organisms like coagulase-negative staphylococci, viridans group streptococci and enterococci [25]. In fact, coagulase-negative staphylococci are the most common blood culture contaminant, typically representing 70-80% of all contaminated blood cultures [26,27].
Conclusion
The presence of identical bacterial strains in both colonies and plasma further suggests that examined samples arrived at the laboratory contaminated. However, the fact that many cancer patients are immunosuppressed and susceptible to the bacteria we investigated makes it more difficult to determine the contamination source. In addition, a lack of information about the clinical condition of patients, blood drawing procedure and laboratory environment in which blood drawing was performed prevents us from making a clear deduction. Therefore, to determine the actual source of contamination, additional studies must be performed with an increased number of samples and improved patient information.
Conflicts of Interest
The authors declare no conflict of interest
19246
References
- MałgorzataMikulskaaClaudioViscoliaChristinaOraschbDavid M.LivermorecDianaAverbuchdCatherineCordonniereMuratAkovafthe Fourth European Conference on Infections in Leukemia Group (ECIL-4), a joint venture of EBMT, EORTC, ICHS, ELN and ESGICH/ESCMID Aetiology and resistance in bacteraemias among adult and paediatric haematology and cancer patients
- Brecher ME, Hay SN (2005) Bacterial contamination of blood components. ClinMicrobiol Rev 18: 195-204.
- Holland T, Fowler VG, Shelburne SA (2014) Invasive gram-positive bacterial infection in cancer patients. Clin Infect Dis 59 Suppl 5: S331-S334.
- Brakstad OG, Aasbakk K, Maeland JA (1992) Detection of Staphylococcus aureus by polymerase chain reaction amplification of the nuc gene. J ClinMicrobiol 30: 1654-1660.
- Liu D, Hollingshead S, Swiatlo E, Lawrence ML, Austin FW (2005) Rapid identification of Streptococcus pyogenes with PCR primers from a putative transcriptional regulator gene. Res Microbiol 156: 564-567.
- Liu D, Swiatlo E, Austin FW, Lawrence ML (2006) Use of a putative transcriptional regulator gene as target for specific identification of Staphylococcus epidermidis. LettApplMicrobiol 43: 325-330.
- Greisen K, Loeffelholz A, Purohit A, Leong D (1993) PCR Primers and Probes for the 16S rRNA Gene of Most Species of Pathogenic Bacteria, Including Bacteria Found in Cerebrospinal Fluid. Journal of clinical microbiology 32: 335-351.
- Osterlund A, Popa R, Nikkila T, Scheynius A, Engstrand L (1997) Intracellular reservoir of Streptococcus pyogenes in vivo: a possible explanation for recurrent pharyngotonsillitis. Laryngoscope 107: 640-647.
- Cleary PP, McLandsborough L, Ikeda L, Cue D, Krawczak J (1998) High-frequency intracellular infection and erythrogenic toxin A expression undergo phase variation in M1 group A streptococci. MolMicrobiol 28: 157-167.
- Ferretti JJ, McShan WM, Ajdic D, Savic DJ, et al. (2001) Complete genome sequence of an M1 strain of Streptococcus pyogenes. ProcNatlAcadSci USA 98: 4658-4663.
- Beres SB, Sylva GL, Barbian KD, Lei B, Hoff JS, et al. (2002) Genome sequence of a serotype M3 strain of group A Streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. ProcNatlAcadSci USA 99: 10078-10083.
- McShan WM, Ferretti JJ, Karasawa T, Suvorov AN, Lin S, et al. (2008) Genome sequence of a nephritogenic and highly transformable M49 strain of Streptococcus pyogenes. J Bacteriol 190: 7773-7785.
- Smoot JC, Barbian KD, Van Gompel JJ, Smoot LM, Chaussee MS, et al. (2002) Genome sequence and comparative microarray analysis of serotype M18 group A Streptococcus strains associated with acute rheumatic fever outbreaks. ProcNatlAcadSci USA 99: 4668-4673.
- Banks DJ, Porcella SF, Barbian KD, Martin JM, Musser JM (2003) Structure and distribution of an unusual chimeric genetic element encoding macrolide resistance in phylogenetically diverse clones of group A Streptococcus. J Infect Dis 188: 1898-1908.
- Green NM, Zhang S, Porcella SF, Nagiec MJ, Barbian KD, et al. (2005) Genome sequence of a serotype M28 strain of group a streptococcus: potential new insights into puerperal sepsis and bacterial disease specificity. J Infect Dis 192: 760-770.
- Beres SB, Musser JM (2007) Contribution of exogenous genetic elements to the group A Streptococcus metagenome. PLoS One 2: e800.
- Holden MT, Scott A, Cherevach I, Chillingworth T, Churcher C, et al. (2007) Complete genome of acute rheumatic fever-associated serotype M5 Streptococcus pyogenes strain manfredo. J Bacteriol 189: 1473-1477.
- Nakagawa I, Kurokawa K, Yamashita A, Nakata M, Tomiyasu Y, et al. (2003) Genome sequence of an M3 strain of Streptococcus pyogenes reveals a large-scale genomic rearrangement in invasive strains and new insights into phage evolution. Genome Res 13:1042-1055.
- Kuroda M, Ohta T, Uchiyama I, Baba T, Yuzawa H, et al. (2001) Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357:1225-40.
- Gill SR, Fouts DE, Archer GL, Mongodin EF, Deboy RT, et al. (2005) Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J Bacteriol 187: 2426-2438.
- Montassier E, Batard E, Gastinne T, Potel G, de La Cochetiere MF (2013) Recent changes in bacteremia in patients with cancer: a systematic review of epidemiology and antibiotic resistance. Eur J ClinMicrobiol Infect Dis 32: 841-850.
- Domenech A, Ardanuy C, Grau I, Calatayud L, Pallares R, et al. (2014)Evolution and genetic diversity of the Spain23F-ST81 clone causing adult invasive pneumococcal disease in Barcelona (1990-2012). J AntimicrobChemother 69: 924-931.
- Shelburne SA, Tarrand J, Rolston KV (2013) Review of streptococcal bloodstream infections at a comprehensive cancer care center, 2000-2011. J Infect 66:136-46.
- Garcia-Vidal C, Ardanuy C, Gudiol C, Cuervo G, Calatayud L, et al. (2012) Clinical and microbiological epidemiology of Streptococcus pneumoniae bacteremia in cancer patients. J Infect 65: 521-527.
- Lujan M, Burgos J, Gallego M, Falco V, Bermudo G, et al. (2013) Effects of immunocompromise and comorbidities on pneumococcal serotypes causing invasive respiratory infection in adults: implications for vaccine strategies. Clin Infect Dis 57: 1722-1730.
- Weinstein MP (2003) Blood culture contamination: persisting problems and partial progress. J ClinMicrobiol 41: 2275-2278.
- Calfee DP, Farr BM (2002) Comparison of four antiseptic preparations for skin in the prevention of contamination of percutaneously drawn blood cultures: a randomized trial. J ClinMicrobiol 40: 1660-1665.







