Aml El-Sayed A1, Ahmed M1*, Abbas Mohamed A2, Mohamed Sayed H3, Hala MA3 and Ahmed F3
1Department of Microbiology, Al-Azhar University, Cairo, Egypt
2Department of Microbiology, Cairo University, Cairo, Egypt
3Department of Internal Medicine, Cairo University, Cairo, Egypt
Corresponding Author:
Ahmed Mora
Department of Biochemistry
Al-Azhar University
Naser City 11884, Cairo, Egypt
Tel: +201069893392
E-mail: ahmedtalaatmora@gmail.com
Received Date: October 08, 2017; Accepted Date: October 16, 2017; Published Date: October 23, 2017
Citation: El-Sayed AA, Ahmed M, Mohamed AA, Sayed HM, Hala MA, et al. (2017) Detection of Helicobacter Pylori Infection in Biliary System in Egyptian Patients with Calcular Obstructive Jaundice. Arch Clin Microbiol. Vol. 8 No. 5:65 doi:10.4172/1989-8436.100065
Copyright: © 2017 El-Sayed AA, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Keywords
Gallstone disease; Helicobacter; Obstructive jaundice-nested PCR
Introduction
Helicobacter pylori (H. pylori), is a gram-negative microaerophilic bacterium found usually in the stomach, was first identified by Warren and Marshall [1] in 1982 and soon after it was linked with chronic gastritis and gastric ulcers [2]. Although H. pylori is recognized as a human pathogen associated with gastric lesions, studies have revealed the presence of Helicobacter species with some extragastric diseases such as cardiovascular diseases, lung diseases, hematologic diseases, eye and skin diseases, hepatobiliary diseases, diabetes mellitus, and neurological disorders [3].
Gallstone disease is one of the common problems affecting the digestive tract however, the cause of gallstone formation beginning with a change in the composition of bile, leading to stones formation [4]. Different studies suggest that bacterial infection has an important role in the formation of brown pigmented gallstones and that the formation of pure cholesterol gall-stones depends mainly on cholesterol saturation and solubility [5].
In many studies, the presences of different Helicobacter species were shown in the hepatobiliary system [4,6]. Bile-resistant hepatic Helicobacter species such as H. bilis, H. pullorum, and F. rappini were isolated from gall bladder (GB) mucosa and bile juice of patients with chronic cholecystitis and gallstones so these agents may be the key elements in the development of various GB-related diseases, especially GB cancer [7].
Diagnosis of H. pylori infection includes invasive (culture of Biopsy specimens taken by endoscopy) and non-invasive (serologic testing of the patient's serum antibody response to the organism) methods [8,9]. Recently Polymerase chain reaction (PCR) has been a reliable and highly sensitive tool for detection of H. pylori gene sequences in clinical specimens [10,11].
Several studies suggest Helicobacter species as an etiological agent in gallstone formation, while another study showed no association [12].
In this study, we aimed to detect specific Helicobacter strains identified by PCR assay uses a nested set of primer in biliary system from Egyptian patients with calcular obstructive jaundice.
Material and Methods
Subjects
This prospective study was conducted from January 2014 to April 2016 after permission was granted from Cairo university ethical committee. Our study included 75 patients referred to El-Ibrashi unit of Gastroenterology and Endoscopy Kaser El-Eini Hospital Cairo University complaining of calcular obstructive jaundice proved with imaging either transabdominal ultrasound or Magnetic resonance cholangiopancreatography (MRCP). All patients undergo ERCP examination who agreed to participate in the study were appointed to the endoscopy room on the day of the procedure.
Informed consent
The patients were 100% compliant to the study. For confidentiality, their names were omitted and replaced by numerical codes.
Clinical specimens
Bile juice was collected by intra-procedure aseptic aspiration using catheter through the inner endoscopic channel for bacterial culture and PCR analysis. Bile samples used for PCR were directly stored at -20°C. Gastric Biopsy specimens were also used for rapid urease test and bacterial culture.
Microbiological culture
Bile juice and part of gastric biopsy were cultured immediately on Columbia blood agar (CM331, Oxoid Ltd, Basingstoke, UK) with Dents H. pylori selective supplement. Plates were incubated under a microaerophilic atmosphere at 37°C for 5-7 days. The microorganism was identified as H. pylori by colony morphology, Gram staining, and positive urease, catalase and oxidase test [13].
During endoscopy, part of gastric biopsy was taken and tested for urease activity by rapid urease test using Kimberrly-Clark, Utah, USA test, the test was read at 2 min, 30 min, 2 h and 24 h, the change in color from a light yellow to pink indicating positive results.
DNA extraction from bile juice
The refrigerated bile samples were pelleted by centrifugation for 10 minutes at 14,000 rpm, then incubated with lysis buffer 10 mMTris-Hcl (pH 8.5), 10 mM EDTA, 100 mM NaCl, 0.5% SDS) and proteinase K at 55°C for 8 hours. Then DNA was extracted twice with phenol-chloroform, and purified using a QIAamp DNA kit (QIAGEN, Hilden, Germany).
Polymerase chain reaction of DNA
The PCR method was used to amplify 16S rRNA of Helicobacter species from the bile samples, the PCR product was re-amplified with nested PCR primers designed by Qiagen-Germany. The sequence of primers for Helicobacter 16s rRNA are (5'-GCTATGACGGGTATCC-3') and (5'-GATTTTACCCCTACACCA-3'). The primers of H.pylori (26 KDa surface antigen) are (5'-TGGCGTGTCATTGACAGCGAGC-3' and (5'-CCTGCTGGGCATACTTCACCATG-3'). All reactions were 50 μL in volume and performed with an automated Gene Amp PCR system 9600 (Perkin Elmer, Norwalk, CT, USA). Reaction mixtures contained 0.2 mM of each oligonucleotide primer, 10x PCR buffer [25 mM TAPS (pH 9.3), 50 mMKcl, 2 mM MgCl2, 1 mM 2-mercaptoethanol], 0.5 U of Taqpolymerase, 0.4 μL of each primer, and 5 μL of template DNA. The PCR products were denatured at 94°C for 1 minute, annealed at 55°C for 1 minute, and elongated at 72°C for 1minute. The PCR reaction ended at 35 cycles. Then, 50μL mixture containing 3 μL of PCR product was reamplified by nested PCR. Final nested PCR products were electrophoretically separated in a 1.5% agarose gel, stained with ethidium bromide, and visualized under UV light in comparison with100 bp molecular weight ladder markers (Gibco BRL).
Results
Culture of H. pylori from gastric biopsy and bile specimens
Our prospective study included 32 men and 43 women with ages ranged from 31 to 63 years (mean age 51.4 years). All patients complaining of calcular obstructive jaundice were proved with imaging modality either trans-abdominal ultrasound or MRCP. Therapeutic ERCP was done and bile juice specimens were obtained during the procedure. Bile juice was collected by aseptic aspiration using catheter through the inner endoscopic channel and bacterial cultures were performed immediately in all patients. Gastric biopsy was taken during the procedure for H. pylori culture and rapid urease testing. Bile samples culture showed no growth for H. pylori however 5 gastric biopsy samples (6.7%) were positive. The rapid urease test in the gastric mucosa was negative in 45 (60%) and positive in 30 (40%) of these patients Table 1.
| Test |
Percentage % |
| Bile juice culture |
0 |
| Gastric biopsy culture |
6.7 |
| Gastric biopsy rapid urease |
40 |
| Helicobacter specific 16S rRNA PCR |
14.7 |
| H pylori26 kDa surface antigen PCR |
14.7 |
Table 1: Percentage of patient's different tests.
Detection of H. pylori using nested PCR assay
Helicobacter specific 16S rRNA PCR product expected at 400-bp were positive for 11(14.7%) out of 75 bile juice samples Figure 1. To identify H. pylori, specific primer pairs HPF and HPR were used to generate amplicons of approximately 298-bp in samples positive for Helicobacter 16S rRNA Figure 2. Patient samples were considered to be positive for H. pylori by PCR amplification if the 400 bp band was seen in the initial reaction and the 298 bp was seen in the internal nested reaction Table 1.
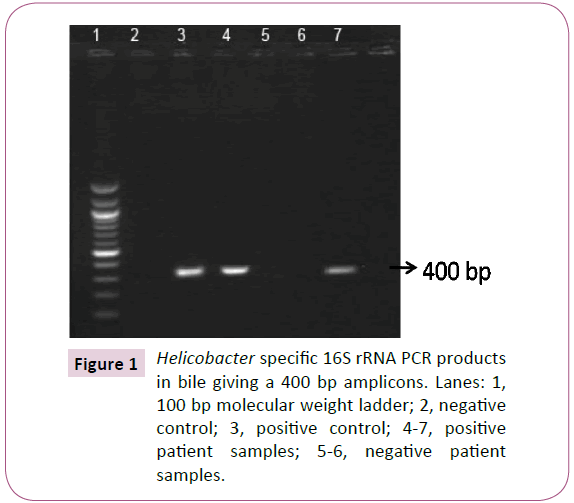
Figure 1: Helicobacter specific 16S rRNA PCR products in bile giving a 400 bp amplicons. Lanes: 1, 100 bp molecular weight ladder; 2, negative control; 3, positive control; 4-7, positive patient samples; 5-6, negative patient samples.
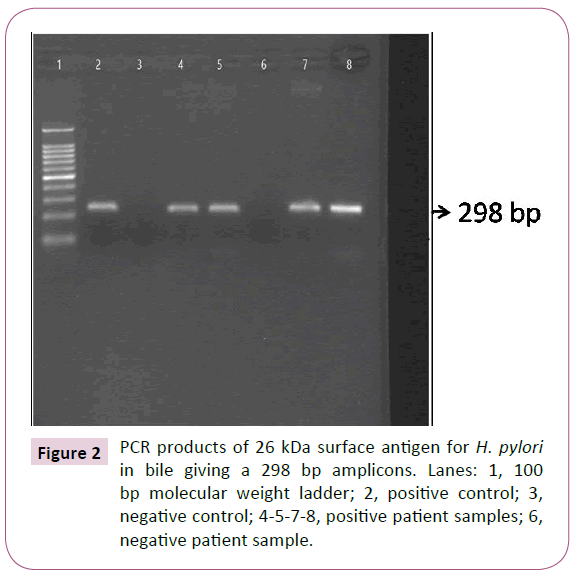
Figure 2: PCR products of 26 kDa surface antigen for H. pylori in bile giving a 298 bp amplicons. Lanes: 1, 100 bp molecular weight ladder; 2, positive control; 3, negative control; 4-5-7-8, positive patient samples; 6, negative patient sample.
Disccusion
Extragastric diseases have been suggested in many studies due to Helicobacter species infection. The Helicobacter species that have implicated as a cause of hepatobiliary diseases are H. pylori, H. rodentium, Flexispira rappini and H. pullorum [14,15].There is evidence that many Helicobacter species such as F. rappini, H. bilis, H. canis, H. hepaticus, H. cholecystus and H. pullorum may be able to survive in bile juice [16,17].
In this study, Helicobacter DNA was detected in 14.7% of the patient bile juice samples. In addition, all bile samples positive for Helicobacter DNA were also positive for H. pylori identified using specific 26 kDa surface antigen primers. In accordance to our result lee et al. 2010 study show that Helicobacter DNA was detected in 6 out of 48 bile samples [18], in contrast to fallone et al. 2003 study which could not detect any Helicobacter DNA in bile samples [11].
Different studies used enzyme-linked immunoabsorbent assay to identify Helicobacter species in bile samples, however this method is not as specific or sensitive in detecting Helicobacter as PCR and may lead to false positive results due to cross-reactivity between different bacterial organisms or proteins [11,19]. Furthermore, Nested PCR against 16s rRNA Heliobacter genus and H. pylori specific surface antigen used in this study is more specific rather than other studies using genes being limited to Helicobacter species such as urease A that may cross-react with other organisms giving false positive test [20,21].
Since Helicobacter species inhabit the gastrointestinal tract, the most likely source of these organisms is ascending infection from the duodenum [22]. Therefore, the detection of H. pylori using rapid urease test was done on the patient's gastric biopsy. 40% of the gastric biopsies were positive for H. pylori urease test including the positive samples for H. pylori in the bile detected by PCR. These results suggesting that bile regurge may have a role to allow surviving of H. pylori that are resistant to bile salts and causing inflammation and stone formation.
Unfortunately, we could not isolate any H. pylori from bile samples in this study and others studies probably because the difficult culture of this fastidious organism and the high sensitivity to the bile salts that may inhibit its viability [23,24].
In summary, our data suggest that bacterial infection of H. pylori may have a role in the formation of gallstones that may be due to cholesterol or calcium precipitation [25]. Future studies focusing in the route of H. pylori infection and the effect of bacterial eradication on the development of gallstone will be needed.
Funding
No funding was required to produce this review article.
Conflict of Interest
The authors declare they have no conflict of interest.
20889
References
- Warren JR, Marshall B (1983) Unidentified curved bacilli on gastric epithelium in active chronic gastritis. (Letter) Lancet 1: 1273-1275.
- Marshall BJ, Warren JR (1984) Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet 1: 1311-1315.
- Suzuki H, Franceschi F, Nishizawa T, Gasbarrini A (2011) Extragastric manifestations of Helicobacter pylori infection. Helicobacter 3: 65-69.
- Attaallah W, Yener N, Ugurlu MU, Manukyan M, Asmaz E, et al. (2013) Gallstones and Concomitant Gastric Helicobacter pylori Infection. Gastroenterol Res Pract 13: 643109.
- Apostolov E, Al-Soud WA, Nilsson I, Kornilovska I, UsenkoV, et al (2005) Helicobacter pylori and other Helicobacter species in gallbladder and liver of patients with chronic cholecystitis detected by immunological and molecular methods. Scand J Gastroenterol 40: 96-102.
- Chu CL, Li YQ, Zhang Y, Li WJ, Zhao XC (2003) Expression of extracellular-signal regulated protein kinase in gastric carcinoma tissues and its relation with Helicobacter pylori infection. ShijieHuarenXiaohuaZazhi 11: 481-482.
- Lin TT, Yeh CT, Wu CS, LiawYF (1995) Detection and partial sequence analysis of Helicobacter pylori DNA in the bile samples. Dig Dis Sci 40: 2214-2219.
- Vaira D, Malfertheiner P, Megraud F, Axon ATR, Deltenre M, et al. (1999) HpSA European Study Group: Diagnosis of Helicobacter pylori infection with a new non-invasive antigen-based assay. Lancet 354: 30-33.
- Clayton CL, Kleanthous H, Coates PJ, Morgan DD, Tabaqchali S (1992) Sensitive detection of Helicobacter pylori by using polymerase chain reaction. J ClinMicrobiol 30: 192-200.
- Fallone CA, Tran S, Semret M, Discepola F, Behr M, et al. (2003) Helicobacter DNA in bile: correlation with hepato-biliary diseases. Aliment PharmacolTher 17: 453-458.
- Messini F (2003) Helicobacter pylori and hepatobiliary diseases. ClinTer 154: 55-56.
- Megraud F (1997) How should Helicobacter pylori infection be diagnosed? Gastroenterology 113: 93-98.
- Ward JM, Anver MR, Haines DC, Benveniste RE (1994) Chronic active hepatitis in mice caused by Helicobacter hepaticus. Am J Pathol 145: 959-968.
- Lin TT, Yeh CT, Wu CS, Liaw YF (1995) Detection and partial sequence analysis of Helicobacter pylori DNA in the bile samples. Dig Dis Sci 40: 2214-2219.
- Fox JG, Yan LL, Dewhirst FE (1995) Helicobacter bilissp. nov., a novel Helicobacter species isolated from bile, livers, and intestines of aged, inbred mice. J ClinMicrobiol 33:445-454.
- Fox JG, Shen Z, Feng F, Dufour JF, Kaplan MM, et al. (1998) Enterohepatic Helicobacter spp. identified from humans with primary biliary cirrhosis. Gut 43Suppl 2: A3.
- Lee JW, Lee DH, Lee JI (2010) Identification of Helicobacter pylori in gallstone, bile, and other hepatobiliary tissues of patients with cholecystitis. Gut Liver 4: 60-67.
- Kornilovska I, Nilsson HO, Al-Soud WA, Nilsson I, Hedenbro J, et al. (2001) Helicobacter pylori and Helicobacter pullorum in patients with cholelithiasis. Gut 49 Suppl 2: 68.
- Labigne A, Cussac V, Courcoux P (1991) Shuttle cloning and nucleotide sequences of Helicobacter pylori genes responsible for urease activity. J Bacteriol 173: 1920-1931.
- Vinette KM, Gibney KM, Proujansky R (2004) Comparison of PCR and clinical laboratory tests for diagnosing H. pylori infection in pediatric patients. BMC Microbiol 4: 5.
- Figura N, Cetta F, Angelico M (1998) Most Helicobacter pylori infected patients have specific antibodies, and some also have H. pylori antigens and genomic material in bile: is it a risk factor for gallstone formation? Dig Dis Sci 43: 854-862.
- Messini F (2003) Helicobacter pylori and hepatobiliary diseases. ClinTer 154: 55-56.
- FarshadSh, Alborzi A, MalekHosseini SA, Oboodi B, Rasouli M, et al. (2004) Identification of Helicobacter pylori DNA in Iranian patients with gallstones. Epidemiol Infect. 132: 1185-1189.
- Belzer C, Kusters JG, Kuipers EJ, Van Vliet AH (2006) Urease induced calcium precipitation by Helicobacter species may initiate gallstone formation. Gut 55: 1678-1679.







