Introduction
Psittacine Beak and Feather Disease (PBFD) is a viral disease that was first discovered in 1975 in cockatoos in Australia [1]. Since then, several outbreaks have been reported in parrots in many countries, including New Zealand [2], South Africa [1], Europe [3], Asia [4-6] and USA [7,8]. The causative agent, BFDV, has a circular ssDNA ambisense genome [9] with two major open reading frames (ORF). ORF V1 encodes for the replication-associated protein (Rep) and ORF C1 encodes for the capsid protein [10]. A third ORF has been identified, but its function remains unknown [8].
The symptoms of chronic PBFD consist of feather loss and abnormalities in feather or beak shape resulting from damage to the epidermis of the feathers and beak [11]. Acquired immunodeficiency as a result of BFDV infection often leads to fatal secondary infections [12].
Several methods have been established to detect BFDV infection, including Haemagglutination (HA) and Haemagglutination Inhibition (HI) assays [13], histology [11], blocking ELISA [14], polymerase chain reaction (PCR) [1,8,15-18], real-time PCR [19] and quantitative real-time PCR [20]. However, these methods are time-consuming and require expensive instruments.
Loop-Mediated Isothermal Amplification (LAMP) is a molecular method that is used to amplify DNA or RNA under isothermal conditions and was developed by Notomi et al. [21]. Several studies suggested that LAMP could be a rapid and highly specific assay that amplifies DNA with high specificity, efficiency and rapidity under isothermal conditions [21-24]. The LAMP reaction is an auto-cycling strand amplification carried out by a Bst DNA polymerase and two pair of specific primers that recognize six sequences on the target DNA. The final LAMP products have stem-loop DNA with various lengths that can be detected by a ladder pattern of bands on a DNA agarose gel or can be visualized as precipitates in a turbid solution with no specific reagent or equipment requirement for the detection of amplified DNA [21,25]. On the other hand, the entire duration of the LAMP assay, including DNA extraction, the reaction, and the final visualization of amplification products, is less than 2.5 h whereas at least 5 h is needed for a conventional PCR-based method [21,26]. In this study, we established a LAMP assay to detect BFDV, for which we determined its sensitivity and specificity.
Materials and Methods
Plasmid and clinical samples
Porcine Circovirus Type 2 (PCV2), pigeon circovirus and Avian Polyoma Virus (APV) specimens were obtained from field and identified by PCR and sequencing. A total of 11 suspected BFDV liver and spleen specimens from different areas of Taiwan were used for diagnostic purposes.
Confirmed clinical samples of PCV2, PiCV and APV were used as controls for specificity experiments. APV is a common virus that causes psittacine species diseases [27-30]. The nucleotide sequence homology in the ORF C1 region between BFDV and PCV2 is approximately 49.1~59% [31]. The recombinant plasmid yT&A-BFDV was used for temperature, time and sensitivity test of LAMP reaction. Eleven suspected PBFD samples were used for clinical sample test of LAMP. A 10% suspension of mixture of liver and spleen from same samples was prepared in PBS (pH7.2) for DNA isolation. All samples were stored at -80°C.
Plasmid extraction
Following the manufacturer’s instruction, plasmid extraction was done using a DNA mini plasmid kit (Qiagen, Hilden, Germany). The purified plasmid was eluted in 60 μL of sterile water and stored at -20°C until later use.
DNA extraction
First, 500 μL of tissue samples that have been frozen and thawed three times was digested by 50 μL of 10% SDS and 10 μL of proteinase K (20 mg/mL) at 55°C for 2 h. Then, DNA was extracted with TRIS saturated phenol, chloroform and absolute ethanol according to the manufacturer’s instruction (Thermo Fisher Scientific, CA, USA). Lastly, DNA was washed with 75% ice-cold ethanol. The precipitated DNA was dissolved in 20 μL of sterile water and stored at -20°C for later use.
LAMP primer design
Forty-one complete genomes, including the 14 subgroups (A to N) from Varsani’s study [35], were used for primer design. A set of four primers was designed according to the conserved region (200-900) of 41 published full length BFDV genome (GQ396652.1, GQ396653.1, GQ396654.1, AF311298.1, AF311302.1, AF311297.1, AY450434.1, AY450435.1, AY450436.1, AF311300.1, AF311301.1, FJ685980.1, FJ685978.1, GU015012.1, GU015013.1, GU015023.1, AF311299.1, FJ685989.1, FJ685970.1, HM748919.1, AY521236.1, DQ397818.1, GU047347.1, GQ120621.1, AY521238.1, FJ685985.1, AF311296.1, AY521235.1, HM748929.1, HM748928.1, HM748927.1, HM748939.1, HM748938.1, HM748918.1, HM748925.1, AY450439.1,AY450438.1, AY450442.1, GQ165757.1, AB277746.1 and AB277747.1). He primers were designed using Primer Explorer ver. (https:// primerexplorer.jp/elamp4.0.0/index.html). A forward primer (LAMP-F3), a backward primer (LAMP-B3), a forward inner primer (LAMP-FIP) and a backward inner primer (LAMP-BIP) were used for the LAMP method. The sequences and binding sites of the primers are shown in Table 1.
| Primer |
Type |
Length |
Sequence |
Product |
Pathogen |
Genome position |
References |
| PBFDV-ORFF |
Forward |
18 |
AACCCTACAGACGGCGAG |
717 |
Psittacine Beak and feather disease virus |
182-898 |
15 |
| PBFDV-ORFR |
Reverse |
20 |
GTCACAGTCCTCCTTGTACC |
|
|
|
|
| PCV2 ORF2F |
Forward |
21 |
ACGTATCCAAGGAGGCGTTAC |
703 |
Porcine Circovirus-type-2 |
1031-1733 |
26 |
| PCV2 ORF2R |
Reverse |
20 |
AGGGTTAAGTGGGGGGTCTT |
|
|
|
|
| PiCV-Pet32aF |
Forward |
27 |
GAATTCCAGATGAGAAGGCGGAGATTC |
820 |
Pigeon Circovirus |
1166-1985 |
26 |
| PiCV-Pet32aR |
Reverse |
31 |
CTCGAGCATCTGCAAAACACTGGTTACAATC |
|
|
|
|
| APV-VP1F |
Forward |
24 |
CTTATGTGGGAGGCTGTCAGTGTT |
550 |
Avain polyomavirus |
2183-2732 |
30 |
| APV-VP1R |
Reverse |
24 |
TACTGAAATAGCGTGGTAGGCCTC |
|
|
|
|
| LAMP-F3 |
Forward Outer |
18 |
TTGTGGCGAGAGACGGTC |
- |
Psittacine Beak and feather disease virus |
462-479 |
This research |
| LAMP-B3 |
Backward Outer |
20 |
CCGTAGATGACGTCAACCTC |
|
|
637-656 |
|
| LAMP-FIP |
Forward Inner |
39 |
AACTCTCGCGCGACTTCCTTCGCTTTCGACGGAGCTGTT |
|
|
459-477+505-524 |
|
| LAMP-BIP |
Forward Inner |
40 |
CATGGGCGGGGCTTGCATAAAGTCTTGAAATCACGTGGGC |
|
|
544-563+589-608 |
|
Table 1: The specific primers for following diseases.
LAMP reaction
The LAMP assay was performed using the Loopamp DNA amplification kit (Eiken Chemical Co. Ltd., Tokyo, Japan). In brief, the assay was performed with the following optimized reaction mixture: 25 μL of a mixture containing 12.5 μL of reaction mix buffer, 1 μL of the template DNA, 40 pmol (each) of primers FIP and BIP, 5 pmol of primers B3 and F3, and 1 μL of Bst DNA polymerase. To determine the optimal conditions for sensitivity and selectivity, the LAMP reactions were performed at a range of temperatures (60°C- 65°C) for different time periods (30 - 60 min). After each incubation time and temperature, reactions were stopped by heating at 80? for 5 min. LAMP products were detected using 2% agarose gel electrophoresis with ethidium bromide staining.
PCR
Three primer pairs (PCV2 ORF2F and PCV2 ORF2R for PCV2; PiCVPet32aF and PiCV-Pet32aR for PiCV; APV-VP1F and APV-VP1R for APV) were used to study the specificity of LAMP when compared with PCR. PCR was carried out in a 25 μL reaction volume containing final concentrations of 0.4 μM of each primer, dNTP mixture (0.2mM each dATP, dCTP, dGTP, dTTP) (Fermentas, CA, USA), 1.5 mM MgCl2 (Fermentas, CA, USA), 2.5 μL of PCR buffer (Fermentas, CA, USA), 1 μL of extracted DNA and 2.5 U of Taq DNA polymerase (Fermentas, CA, USA). The PCR conditions were as follows: an initial activation at 95°C for 10 min, and then 35 cycles of amplification (3 min at 95°C, 45 s at 53°C, 30 s at 72°C). The PCR products were analyzed by 1.5% agarose gel electrophoresis.
Sensitivity and specificity of LAMP
The BFDV template was diluted serially 10-fold in double-distilled water (diluted from 3.5 pg to 3.5 fg) to determine the detection limit. To compare the sensitivity of LAMP with that of PCR, both LAMP (LAMP-F3, LAMP-B3, LAMP-FIP and LAMP-BIP primer pairs) and PCR (PBFDV-ORFF and PBFDV-ORFR primer pair) were carried out under optimized condition with. The final optimized condition of LAMP was 63°C for 60 min. DNA of PCV2 (PCV2 ORF2F and PCV2 ORF2R primer pair), PiCV (PiCV-Pet32aF and PiCV-Pet32aR primer pair) and APV (APV-VP1F and APV-VP1R primer pair) were examined to assess the specificity of LAMP and conventional PCR. The sequences of the primers are shown in Table 1. Furthermore, 11 clinical samples were tested simultaneously by LAMP and PCR. Sterile water was used as negative control.
Clinical specimens
During the period 2012-2013, 11 clinical specimens of suspected PBFD samples were collected from different parrot farms in southern Taiwan. A 10% suspension of mixture of liver and spleen from the same sample was prepared in PBS (pH 7.2) for DNA isolation. All samples were stored at −80°C. The use of animals in this research complies with the Taiwan animal welfare law.
Results
Optimization of BFDV LAMP assay
A one-step LAMP assay for the rapid detection of BFDV was developed using a set of 4 primers designed based on highly conserved regions of BFDV. Plasmid yT&A-BFDV was used to optimize the assay reaction temperature and time. In the first step, the LAMP reaction was carried out for 60 min at 60, 61, 62, 63, 64, and 65°C. As shown in Figure 1a, ladder pattern of products were generated at 60-65°C. Further experiments showed that there was significant amplification with reaction times for 30, 40, 50 and 60 min at 63°C, yielding ladder pattern of bands on a DNA agarose gel (Figure 1b); thus, 60 min at 63°C was used as the optimal reaction conditions in subsequent assays.
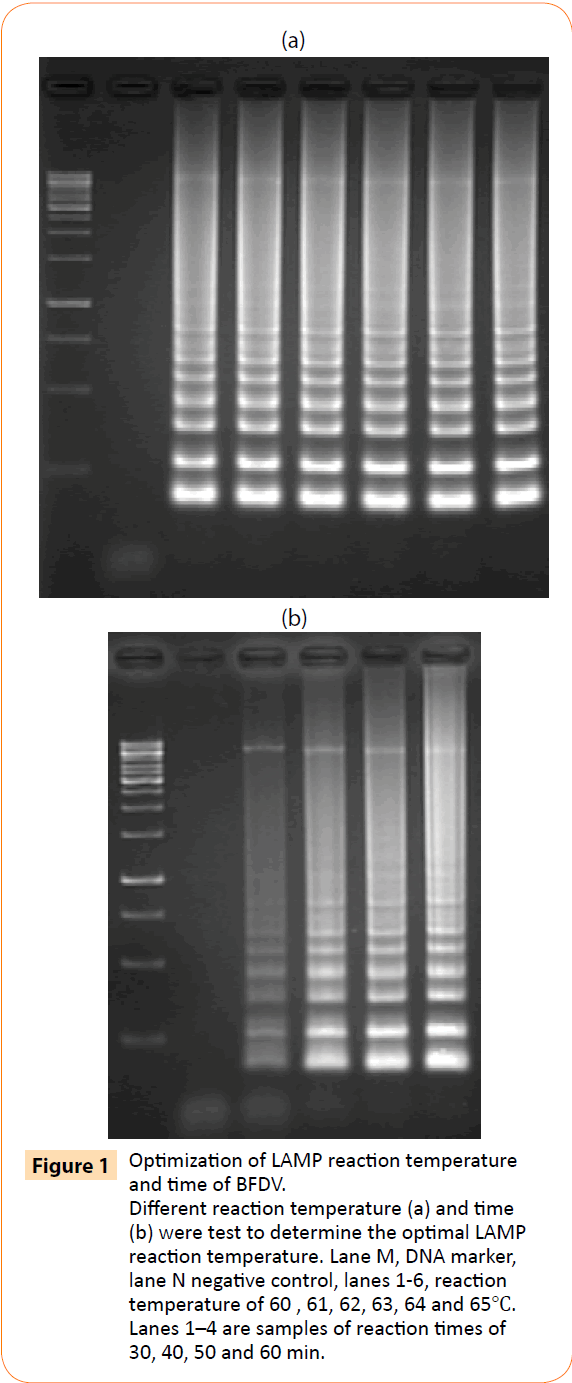
Figure 1: Optimization of LAMP reaction temperature and time of BFDV. Different reaction temperature (a) and time (b) were test to determine the optimal LAMP reaction temperature. Lane M, DNA marker, lane N negative control, lanes 1-6, reaction temperature of 60 , 61, 62, 63, 64 and 65?. Lanes 1–4 are samples of reaction times of 30, 40, 50 and 60 min.
Sensitivity of LAMP and PCR for BFDV
Using serially diluted DNA from recombinant plasmid as template from 3.5 pg to 3.5 fg, LAMP assay and conventional PCR for PiCV were performed to compare their detection limits. All amplicons were analyzed by agarose gel electrophoresis. The detection limit of LAMP (Figure 2b) was 3.5 fg/μL of DNA versus 3.5 fg/μL for conventional PCR (Figure 2a). Thus, the LAMP assay was as sensitive as conventional PCR.
Figure 2: Sensitivity of BFDV detection by PCR (a) and LAMP (b) assay. A 10-fold serial dilution of BFDV DNA was used to determine the sensitivity of the LAMP assay. Lane M, DNA marker; lane N, negative control, lanes 1–5, amplification products obtained with DNA templates of 3.5 pg ~ 3.5 fg
Specificity of LAMP for BFDV
To determine the specificity of the LAMP assay, PiCV, PCV2 and avian polyoma virus samples were tested for cross-reactivity. Viral genomic DNA was used as template for LAMP and conventional PCR detection. By agarose gel electrophoresis, only BFDV gave a positive reaction—a ladder-like pattern of bands (Figure 3b). Moreover, PCR was positive with specific primers for PiCV, PCV2 and avian polyoma virus (Figure 3a). Thus, the LAMP assay was specific for BFDV. Four primers were needed to recognize 6 distinct regions in the LAMP reaction to amplify the target DNA with high specificity.
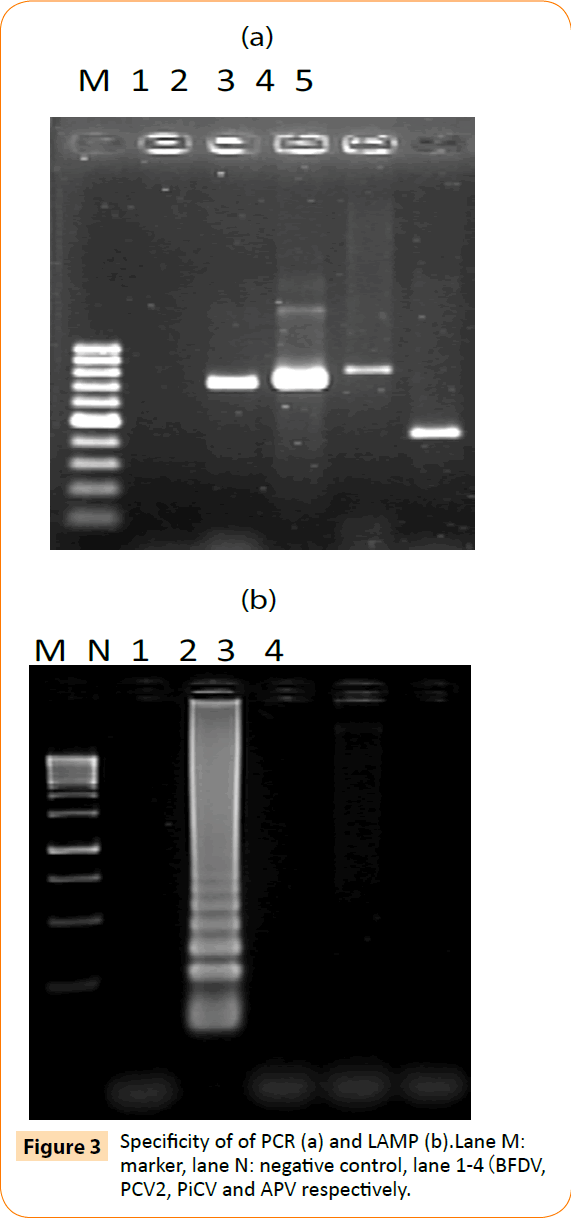
Figure 3: Specificity of of PCR (a) and LAMP (b).Lane M: marker, lane N: negative control, lane 1-4(BFDV, PCV2, PiCV and APV respectively.
LAMP of clinical samples
To confirm its applicability for detecting BFDV in field-obtained samples, the LAMP assay was used to detect BFDV in 11 liver and spleen mixture samples from parrots. All 4 (4/11; 36.3%) samples were positive by LAMP assay, and 7(7/11; 63.6%) samples were negative (Fig 4b). To validate these results, PCR was performed on the same samples. Only 2 (2/11; 18.1%) samples were positive by conventional PCR assay, and 9 (9/11; 81.8%) samples were negative (Figure 4a). These results suggest that the BFDV LAMP assay is more sensitive than PCR in detecting clinical PBFD samples.
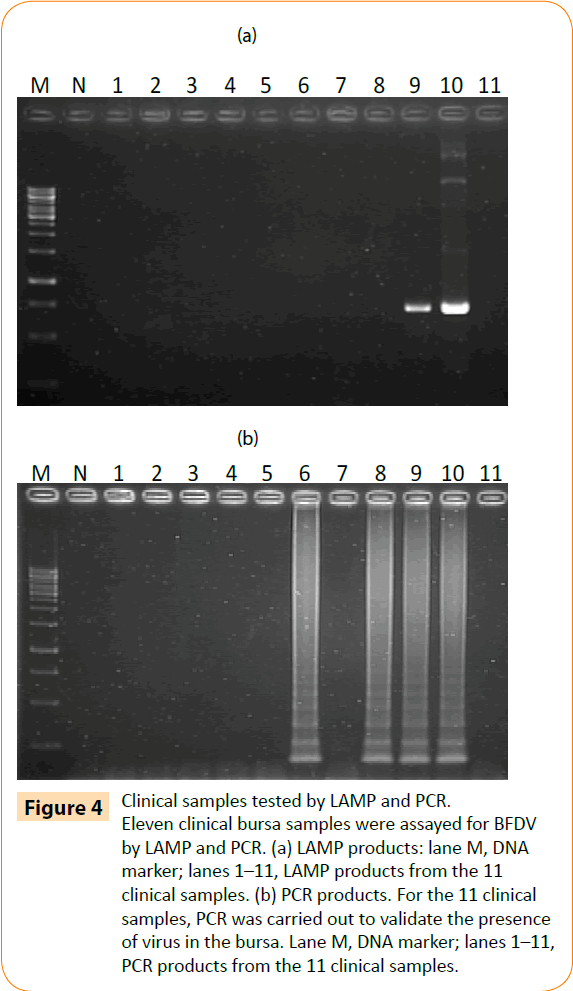
Figure 4: Clinical samples tested by LAMP and PCR. Eleven clinical bursa samples were assayed for BFDV by LAMP and PCR. (a) LAMP products: lane M, DNA marker; lanes 1–11, LAMP products from the 11 clinical samples. (b) PCR products. For the 11 clinical samples, PCR was carried out to validate the presence of virus in the bursa. Lane M, DNA marker; lanes 1–11, PCR products from the 11 clinical samples.
Discussion
BFDV is a significant viral pathogen in parrot species, causing feather loss and abnormalities in feather or beak shape resulting from damage to the epidermis of the feathers and beak [11]. Acquired immunodeficiency as a result of BFDV infection often leads to fatal secondary infections [12]. Amplification methods that are based on PCR, such as conventional PCR and real-time PCR, have been used to detect and quantify this virus [32-34], but application of the recently developed LAMP assay for rapid detection of BFDV has not been reported. As described for many viral pathogens [26], the LAMP assay shows good sensitivity in detecting viruses. The entire duration of the LAMP assay, including DNA extraction, the reaction, and the final visualization of amplification products, is less than 2.5 h whereas at least 5 h is needed for a conventional PCR-based method [26]. Thus, LAMP is a more rapid method of detecting BFDV than PCR. In addition, LAMP can be performed with common and inexpensive equipment. The LAMP reaction is executed in a single tube and only requires a water bath to provide a constant temperature; moreover, the amplification products can be detected by the naked eye. Further still, there are several studies reporting on a direct LAMP assay: by a simple boiling and chilling treatment, no nucleic acid extraction was necessary for the detection of EV71 in nasopharyngeal swab specimens, alpha herpesvirus in skin infection, canine parvovirus in suspected faecal samples, and Mycobacterium tuberculosis in L-J media cultured bacteria [35-38]. It means that on-site diagnosis using LAMP is possible. Based on these advantages, the LAMP assay is considerably more applicable than PCR, particularly in the field.
The optimal conditions for the LAMP reaction in detecting BFDV DNA were 63°C and 60 min. In this study, the limit of detection of LAMP for BFDV was 3.5 fg of viral DNA, suggesting that the LAMP assay is as sensitive as a PCR-based assay. In detecting BFDV from experimental and clinical samples, the LAMP method was as least as sensitive as PCR. Also, the specificity of the LAMP reaction was extremely high because it employs 4 primers that recognized 6 distinct regions in the target DNA. In this study, other pathogens, including PCV2, PiCV, and avian polyoma virus, were not detected by LAMP, indicating the high specificity of this assay for BFDV.
Our LAMP assay detected BFDV not only from template DNA but also from tissues of infected parrots, indicating the feasibility of rapid diagnosis in early infection. The final LAMP products have stem-loop DNA with various lengths that can be detected by a ladder pattern of bands on a DNA agarose gel or can be visualized as precipitates in a turbid solution with no specific reagent or equipment requirement for the detection of amplified DNA [21,25]. On the other hand, the entire duration of the LAMP assay, including DNA extraction, the reaction, and the final visualization of amplification products, is less than 2.5 h whereas at least 5 h is needed for a conventional PCR-based method [21,26]. The assay is simple, has high sensitivity and specificity, and is costeffective as it does not require special reagents or sophisticated equipment. The LAMP assay may be useful for rapid diagnosis in the early stages of BFDV infection in parrots, accelerating implementation of disease control protocols.
Conclusion
This is the first report indicating that LAMP is a valuable, rapid method of detecting BFDV with high sensitivity and specificity.
Acknowledgements
These studies were supported by Grants to Kuo Pin Chuang from the National Science Council (NSC) and Council of Agriculture.
Conflict of interest
All authors have no financial or personal relationships with other people or organizations that could inappropriately influence or bias their work.
7771
References
- Albertyn,Tajbhai KM, Bragg RR (2004) Psittacine beak and feather disease virus in budgerigars and ring-neck parakeets in South Africa. Onderstepoort J Vet Res 71: 29-34.
- Massaro M, Ortiz-Catedral L, Julian L, Galbraith JA, Kurenbach B, et al. (2012) Molecular characterisation of beak and feather disease virus (BFDV) in New Zealand and its implications for managing an infectious disease. Arch Virol 157: 1651-1663.
- Julian L, Piasecki T, Chrzastek K, Walters M, Muhire B, et al. (2013) Extensive recombination detected among beak and feather disease virus isolates from breeding facilities in Poland. J Gen Virol 94: 1086-1095.
- Sariya L, Prompiram P, Khocharin W, Tangsudjai S, Phonarknguen R, et al. (2011) Genetic analysis of beak and feather disease virus isolated from captivepsittacine birds in Thailand. Southeast Asian J Trop Med Public Health 42: 851-858.
- Zhuang Q, Chen J, Mushtaq MH, Chen J, Liu S, et al. (2012) Prevalence and genetic characterization of avian polyomavirus and psittacine beak and feather disease virus isolated from budgerigars in Mainland China. Arch Virol 157: 53-61.
- Hsu CM, Ko CY, Tsaia HJ (2006) Detection and sequence analysis of avian polyomavirus and psittacine beak and feather disease virus from psittacine birds in Taiwan. Avian Dis 50: 348-353.
- Olsen G, Speer B (2009) Laboratory reporting accuracy of polymerase chain reaction testing for psittacine beak and feather disease virus. J Avian Med Surg 23: 194-198.
- deKloet E, de Kloet SR (2004) Analysis of the beak and feather disease viral genome indicates the existence of several genotypes which have a complex psittacine host specificity. Arch Virol 149: 2393-2412.
- Bassami MR, Ypelaar I, Berryman D, Wilcox GE, Raidal SR (2001) Genetic diversity of beak and feather disease virus detected in psittacine species in Australia. Virology 279: 392-400.
- Todd D, McNulty MS, Adair BM, Allan GM (2001) Animal circoviruses. Adv Virus Res 57: 1-70.
- Pass DA, Perry RA (1984) The pathology of psittacine beak and feather disease. Aust Vet J 61: 69-74.
- Latimer KS, Rakich PM, Steffens WL, Kircher IM, Ritchie BW, et al. (1991) A novel DNA virus associated with feather inclusions in psittacine beak and feather disease. Vet Pathol 28: 300-304.
- Raidal SR, Cross GM (1994) Thehaemagglutination spectrum of psittacinebeak and feather disease virus.Avian Pathol 23: 621-630.
- Shearer PL, Sharp M, Bonne N, Clark P, Raidal SR (2009) A blocking ELISA for the detection of antibodies to psittacine beak and feather disease virus (BFDV). J Virol Methods 158: 136-140.
- Ypelaar I, Bassami MR, Wilcox GE, Raidal SR (1999) A universal polymerase chain reaction for the detection of psittacine beak and feather disease virus. Vet Microbiol 68: 141-148.
- Kondiah K, Albertyn J, Bragg RR (2006) Genetic diversity of the Rep gene of beak and feather disease virus in South Africa. Arch Virol 151: 2539-2545.
- Ogawa H, Yamaguchi T, Fukushi H(2005) Duplex shuttle PCR for differential diagnosis of budgerigar fledgling disease and psittacine beak and feather disease. MicrobiolImmunol49:227-237.
- Ritchie PA, Anderson IL, Lambert DM (2003) Evidence for specificity of psittacine beak and feather disease viruses among avian hosts. Virology 306: 109-115.
- Raue R, Johne R, Crosta L, Burkle M, Gerlach H, et al. (2004) Nucleotide sequence analysis of a C1 gene fragment of psittacine beak and feather disease virus amplified by real-time polymerase chain reaction indicates a possible existence of genotypes. Avian Pathol 33: 41-50
- Shearer PL, Sharp M, Bonne N, Clark P, Raidal SR (2009) A quantitative, real-time polymerase chain reaction assay for beak and feather disease virus. J Virol Methods 159: 98-104.
- Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, et al. (2000) Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 28: E63.
- Song C, Wan H, Yu S, Han X, Qiu X, et al. (2012) Rapid detection of duck hepatitis virus type-1 by reverse transcription loop-mediated isothermal amplification. J Virol Methods 182: 76-81.
- Wang C, Pang VF, Jeng CR, Lee F, Huang YW, et al. (2011) Detection of porcine circovirus type 1 in commercial porcine vaccines by loop-mediated isothermal amplification. Folia Microbiol 56: 483-489.
- Zhou S, Han S, Shi J, Wu J, Yuan X, et al. (2011) Loop-mediated isothermal amplification for detection of porcine circovirus type 2. Virol J 8: 497.
- Kil EJ, Kim S, Lee YJ, Kang EH, Lee M, et al. (2015) Advanced loop-mediated isothermal amplification method for sensitive and specific detection of Tomato chlorosis virus using a uracil DNA glycosylase to control carry-over contamination. J Virol Methods 213:68-74.
- Tsai SS, Chang YL, Huang YL, Liu HJ, Ke GM, et al. (2014) Development of a loop-mediated isothermal amplification method for rapid detection of pigeon circovirus. Arch Virol 159: 921-926.
- van den Brand JM, Manvell R, Paul G, Kik MJ, Dorrestein GM (2007) Reovirus infections associated with high mortality in psittaciformes in The Netherlands. Avian Pathol 36: 293-299.
- Conzo G, Magnino S, Sironi G, Lavazza A, Vigo PG, et al. (2001) Reovirus infection in two species of Psittaciformes recently imported into Italy. Avian Pathol 30: 43-47.
- Sanchez-Cordon PJ, Hervas J, Chacon de Lara F, Jahn J, Salguero FJ, et al. (2002) Reovirus infection in psittacine birds (Psittacuserithacus): morphologic and immunohistochemical study. Avian Dis 46:485-492.
- Phalen DN, Wilson VG, Gaskin JM, Derr JN, Graham DL (1999) Genetic diversity in twenty variants of the avian polyomavirus. Avian Dis 43: 207-218.
- Todd D, Weston JH, Soike D, Smyth JA (2001) Genome sequence determinations and analyses of novel circoviruses from goose and pigeon. Virology 286: 354-362.
- Duchatel JP, Todd D, Smyth JA, Bustin JC, Vindevogel H (2006) Observations on detection, excretion and transmission of pigeon circovirus in adult, young and embryonic pigeons. Avian Pathol 35: 30-34.
- Duchatel JP, Todd D, Willeman C, Losson B (2009) Quantification of pigeon circovirus in serum, blood, semen and different tissues of naturally infected pigeons using a real-time polymerase chain reaction. Avian Pathol 38: 143-148.
- Franciosini MP, Fringuelli E, Tarhuni O, Guelfi G, Todd D, et al. (2005) Development of a polymerase chain reaction-based in vivo method in the diagnosis of subclinical pigeon circovirus infection. Avian Dis 49: 340-343.
- Mukhopadhyay HK, Amsaveni S, Matta SL, Antony PX, Thanislass J, et al. (2012) Development and evaluation of loop-mediated isothermal amplification assay for rapid and sensitive detection of canine parvovirus DNA directly in faecal specimens.LettApplMicrobiol 55:202-209.
- Nie K, Qi SX, Zhang Y, Luo L, Xie Y, et al. (2012) Evaluation of a direct reverse transcription loop-mediated isothermal amplification method without RNA extraction for the detection of human enterovirus 71 subgenotype C4 in nasopharyngeal swab specimens. PLoS One7:e52486.
- Phetsuksiri B , Rudeeaneksin J, Srisungngam S, Bunchoo S, Roienthong D, et al. (2013) Applicability of in-house loop-mediated isothermal amplification for rapid identification of Mycobacterium tuberculosis complex grown on solid media. Jpn J Infect Dis 66: 249-251.
- Kobayashi T, Yagami A, Suzuki K, Ihira M, Yoshikawa T, et al. (2013) Clinical utility of loop-mediated isothermal amplification assay for the diagnosis of common alpha herpesvirus skin infections. J Dermatol4:1346-8138.









