Keywords
Exosomes; Direct isolation; Flow cytometry; Western blotting; Immunoassay; Magnetic beads; CD9; CD81
Introduction
Exosomes (50-150 nm) are extracellular vesicular structures secreted by cells in culture and found in body fluids [1]. Exosomes are involved in different biological processes including antigen presentation [2], apoptosis, angiogenesis, inflammation and coagulation [3]. They can activate signalling pathways or deliver nucleic acids to distant cells [4-6]. Tumor-derived exosomes can enhance cancer progression, suppressing the immune response and transfer oncogens from tumor host cells [7]. Targeting exosomes could be advantageous in order to increase the efficacy of therapeutic antibodies.
Exosomes, formed by invagination of the endosomal membrane forming multivesicular bodies contain miRNA and mRNA [6,8- 11]. The protein composition mirrors their endocytic origin, MVB formation machinery, tetraspanins (e.g. CD9, CD81), heat shock proteins in addition to lipid-related proteins and phospholipases [12,13] and their possible role in intercellular communication [14].
Currently, exosomes are isolated by differential ultracentrifugation, density gradients or cushions [15], size exclusion [16] or precipitation [17-19]. To obtain ultrapure exosomes or isolation of potential subpopulation of exosomes, an immunomagnetic isolation strategy can be applied by targeting exosomal markers [20,21].
The aim of this study was to address critical factors for direct exosome isolation and to implement this knowledge to establish an easy, reliable and direct method for exosome characterization ensuring compatibility with downstream analysis.
Materials and Methods
Reagents
For isolation of CD9+ and CD81+ exosomes Dynabeads® coated with monoclonal anti-human CD81 (Thermo Fisher Scientific, Oslo, Norway) were applied. Primary antibodies for Western blotting analysis: mouse anti-CD81 (10630D) and CD9 (10626D, Thermo Fisher Scientific, Oslo, Norway). Primary antibodies for flow cytometry: mouse anti-human CD9 PE (555372) and -CD81 PE (555676, BD Biosciences USA). HRP-coupled anti-mouse IgG Trueblot was purchased from Rockland (US). AFP-substrate: DynaLight Substrate with Rapid Glow (4475406), Life Technologies. Chemiluminescence reader: BioTek Synergy (BioRad). AFPconjugated antibodies (CD9 and CD81) for immunoassay (in house conjugation). Protein-A gold was obtained from CMC (Utrecht, Netherlands).
Cell culture
The SW480 cell line (ATCC) was cultured to confluence in RPMI 1640 (10% FCS, 1 mM Sodium-pyruvat) in cell culture bottles (37°C and 5% CO2) and medium was replaced with 50 mL fresh medium. Jurkat cells (ATCC) were seeded at a density of 0.4 x 106 cells per mL in fresh RPMI medium and grown for 3 days before harvest.
Exosome harvest and pre-enrichment
The cell culture medium was replaced with fresh complete medium with FBS 3 days before exosome harvesting. Jurkat and SW480 cells (5 x 107) were cultured for each exosome sample preparation. After 24 h, cells and debris were removed by centrifugation at 350x g for 10 minutes and 2,000 x g for 30 min. For ultracentrifugation, the samples were pre-enriched at 100.000 x g for 70 min at 4°C using a Ti45 rotor (Beckman) and Beckman ultracentrifugation tubes (355622) on a Beckman Optima XPN-100. For precipitation, Total Exosome Isolation kit (Thermo Fisher Scientific, USA) was added to the celland debris-free cell medium (1:2 with exosome isolation reagent and cell medium, respectively). Cell medium and the exosome isolation reagent were mixed by brief vortexing and incubated at 4°C overnight before centrifugated at 4 °C at 10,000 x g for 1 h. The pellet containing pre-enriched exosomes was resuspended in PBS (e.g. 400 μl for 10 ml cell media input).
Size measurement of exosomes
Analysis of pre-enriched exosomes was performed as previously described [22] using the NanoSight LM10 instrument.
Real time quantitative PCR
RNA isolation, reverse transcription and real time quantitative PCR analysis were performed as described earlier [23] using the following primers: Hs99999905_m1 (GAPDH), Hs03023880_g1 (ACTB), Hs99999901_s1 (18S), Let7a: has-let-7a, 000377, and has-miR-16, 000391 (miR16).
Immunolabeling and negative stain of exosomes for electron microscopy
Immunolabeling followed by negative stain and electron microscopy was carried out targeting CD81 as previously described [24] and analyzed at the University of Oslo, Norway on Philips CD100. Images were recorded at 80 kV. Further image processing was done with Adobe Photoshop software.
Preparation of pre-enriched exosomes for Western immunoblotting analysis
The total pre-enriched exosome samples were prepared for Western blotting as described by Oksvold et al [22].
Direct capture of exosomes from cell culture supernatant for flow cytometry, Western blotting analysis and immunoassay
20 μL of magnetic beads (1 x 107 beads/mL 1.3x108 beads/mL, and 0.5 x 107 beads/mL were used for flow cytometry, western blotting and immunoassay, respectively) and washed in 200 μL PBS (0.1 % BSA, 0.2 μm filtered). 0-100 μL of cell-and debris-free cell medium containing exosomes were added to the magnetic beads and incubated for 16- 20 h at 4oC while rotating/mixing unless other incubation times are indicated. After incubation, the tubes containing magnetic beads coated with exosomes were subjected to a short centrifugation followed by applying a magnetic field. Washing the exosome coated beads twice were done by removing the supernatant while being on the magnet and replaced the supernatant with 0.3 mL of PBS (0.1 % BSA). Samples were then processed for flow cytometry or Western blotting. For immunoassay washing was performed two to three times in microtitre plates in 100 μL TBS (0.1% Tween-20) buffer.
Preparation of immunoisolated exosomes for Western immunoblotting analysis
Magnetic beads coated with anti-human CD9 or CD81 were mixed with exosome samples and incubated over night at 4°C while mixing. The beads were washed in PBS (0.1% BSA) and lysed in 25 μl RIPA buffer (150 mM sodium chloride, 1.0% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris, pH 8.0) with proteinase inhibitors followed by 15 min incubation on ice. The lysate was mixed 1:1 with 2x Tris lysis buffer under non-reducing conditions. Samples were further processed for SDS-PAGE.
Flow cytometry
CD9 or CD81 positive exosomes captured directly from cell culture media or after pre-enrichment were resuspended in 300 μl PBS with 0.1% BSA. 100 μl was used per flow staining and incubated for 45-60 minutes on a shaker (1050 rpm) at room temperature. The labelled exosomes were washed and analysed using BD LSRII and BD FACS Diva.
Immunoassay
CD9 (also positive for CD81) positive exosomes captured directly from cell culture media with magnetic beads were stained with AP conjugated CD81 or CD9 antibodies (1:1000 in TBS-T) for 60 minutes on a shaker (1050 rpm) at room temperature. The labelled exosomes were washed three times, transferred to new wells, added substrate and analysed using BioTek Synergy (BioRad) chemiluminescense reader.
Results
Analysis of exosome markers in pre-enriched exosome samples
Prior to analysis, the RNA content of the exosomes was verified. SW480 cell culture media was harvested after 3 days followed by precipitation and analysis of mRNA/rRNA and microRNA content. The exosomes contained full length mRNA/rRNA (GAPDH and 18S) and microRNAs (ACTb, Let7a and miR16) typical for exosomes (Figure 1A). Then, the size distribution was estimated. The precipitated exosomes from SW480 cell culture media was analysed by the Nanosight LM10 instrument. The size distribution showed a size range (top at 111 nm) (Figure 1B) comparable to standard exosome preparations by ultracentrifugation. Finally, the pre-enriched vesicles were identified by electron microscopy. Pre-enriched exosomes were prepared from SW480 cells. Here antigens located on the surface of the exosomes can bind detection antibodies such as the tetraspanin protein CD81, commonly identified in exosomal preparations [25]. Immunoelectron microscopy confirmed the identity of the vesicles both in terms of size and expression of CD81 (Figure 1C and 1D).
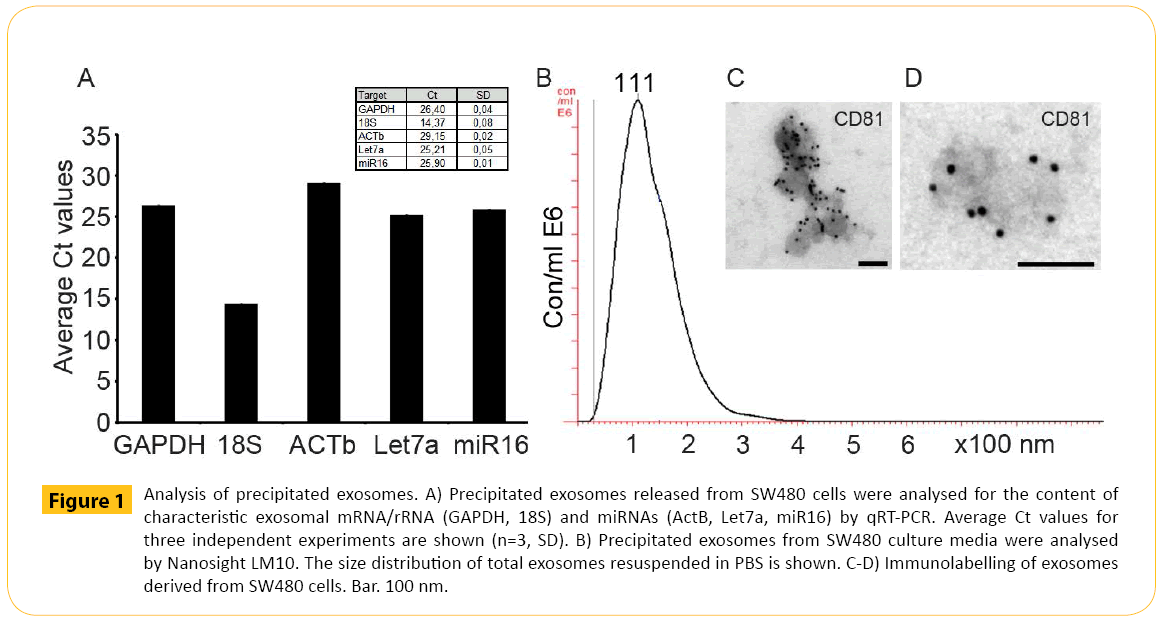
Figure 1: Analysis of precipitated exosomes. A) Precipitated exosomes released from SW480 cells were analysed for the content of characteristic exosomal mRNA/rRNA (GAPDH, 18S) and miRNAs (ActB, Let7a, miR16) by qRT-PCR. Average Ct values for three independent experiments are shown (n=3, SD). B) Precipitated exosomes from SW480 culture media were analysed by Nanosight LM10. The size distribution of total exosomes resuspended in PBS is shown. C-D) Immunolabelling of exosomes derived from SW480 cells. Bar. 100 nm.
Pre-enriched exosomes were then subjected to Western blotting analysis to target the exosomal markers CD9 and CD81. Currently, the most common method for exosome pre-enrichment is differential ultracentrifugation. Therefore, the presence of the exosomal marker CD9 and CD81 in exosomes prepared by ultracentrifugation were compared with CD9 and CD81 expression in exosomes prepared by precipitation (Figure 2A and 2B). Exosomes were pre-enriched from the same cell culture medium using the same amount of input volume. In addition, the same volume of exosomes applied to each well was the same. Therefore, the results from these two pre-enrichment methods should be comparable. The data showed equal amounts of CD9 from SW480 derived exosomes using the two methods for exosome pre-enrichment (Figure 2A). Furthermore, CD81 was also detected in SW480 derived exosomes after pre-enrichment using precipitation (Figure 2A). CD81 was detected in exosomes derived from Jurkat pre-enriched by ultracentrifugation and precipitation. CD9 was absent in equal to the findings by Oksvold et al. [22]. The presence of contaminating non-exosomal vesicles were examined by staining for gp96, an endoplasmic reticulum marker. Gp96 was only observed in cell lysate and absent in preenriched exosomes (data not shown).
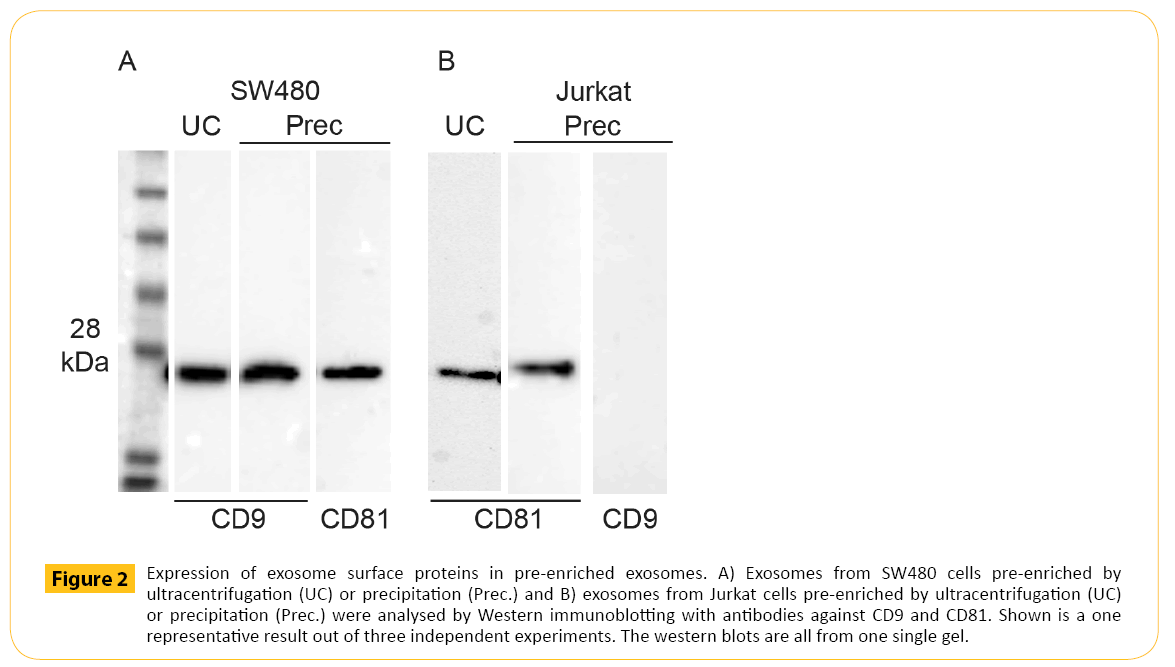
Figure 2: Expression of exosome surface proteins in pre-enriched exosomes. A) Exosomes from SW480 cells pre-enriched by ultracentrifugation (UC) or precipitation (Prec.) and B) exosomes from Jurkat cells pre-enriched by ultracentrifugation (UC) or precipitation (Prec.) were analysed by Western immunoblotting with antibodies against CD9 and CD81. Shown is a one representative result out of three independent experiments. The western blots are all from one single gel.
Analysis of pre-enriched exosome sub-population
Common methods for exosome sample preparation are ultracentrifugation and precipitation. Both methods are challenging in terms of lack of vesicle specificity obtaining vesicle populations with heterogeneous size distributions indicating that other vesicles are co-enriched [26]. Size of exosomes will provide little information on FACS given the limited resolution. Magnetic beads can provide a solid support at a size detectable by flow cytometry. Increased sensitivity and removal of potential protein aggregates and possibly isolate subpopulations of exosomes were obtained by immunomagnetic separation strategy followed by downstream flow analysis. The forward/side scatter plot demonstrated how the magnetic beads were displayed in flow cytometry (Figure 3A). Immunomagnetic isolation of exosomes and staining with isotypic antibody demonstrating the background (Figure 3B-3E, grey histograms) compared to specific signal obtained by relevant CD9 and CD81 staining of bead isolated exosomes (Figure 3B-3E, red histograms). With the purpose of maximize the flow signal and be able to compare data the amount of magnetic beads and the isolation volume were kept constant, ensuring equal binding kinetics. Pre-enriched CD9+ SW480 derived exosomes were isolated using magnetic beads coated with anti-human CD9 antibodies followed by CD9 staining (Figure 3B). The data demonstrated significant staining of CD9. Isolation of CD81+ exosomes from SW480 cells showed the presence of CD81 but at lower signal (Figure 3C). Likewise, pre-enriched Jurkat derived exosomes confirmed the presence of CD81 (Figure 3E). Finally, isolation of CD9+ exosomes from Jurkat cells resulted in signal equivalent to background (Figure 3D).
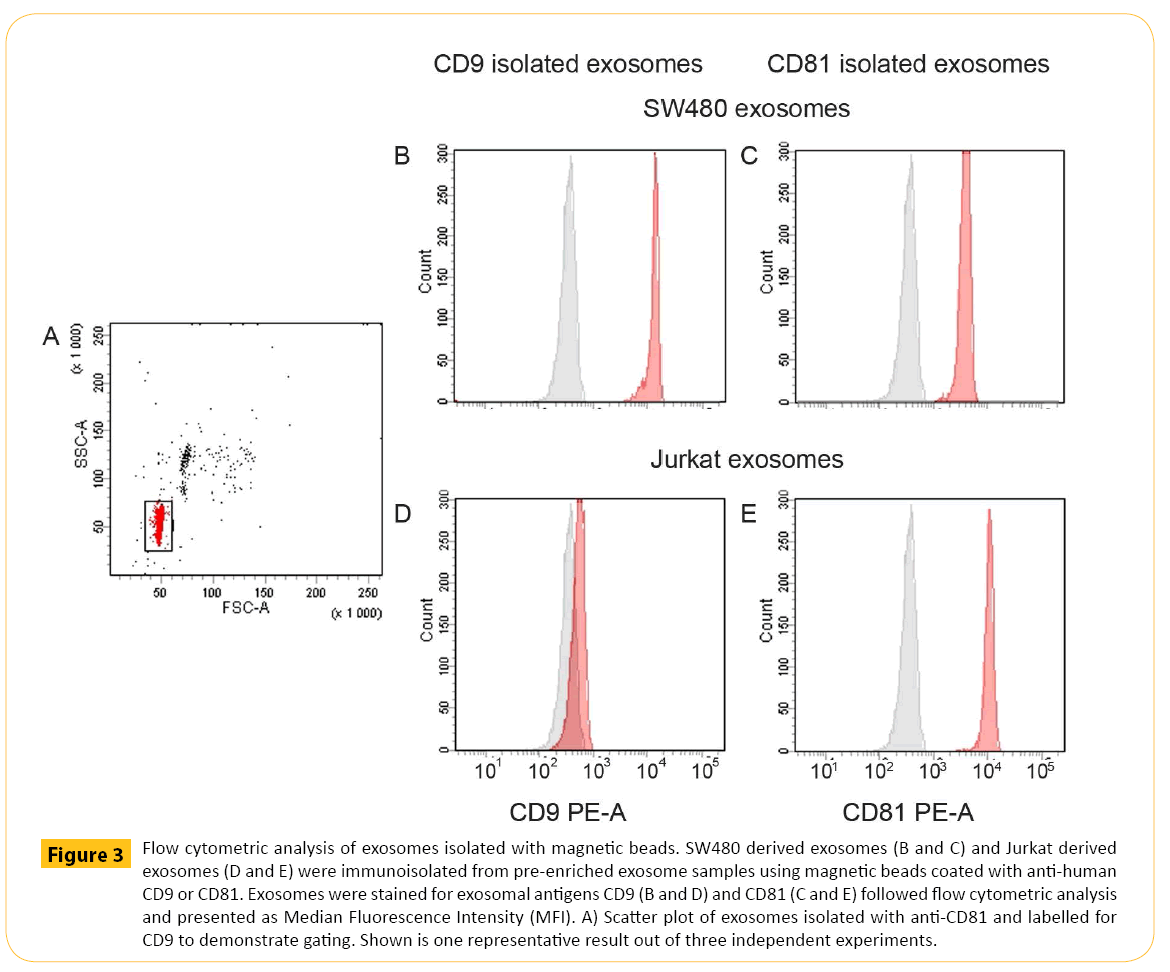
Figure 3: Flow cytometric analysis of exosomes isolated with magnetic beads. SW480 derived exosomes (B and C) and Jurkat derived exosomes (D and E) were immunoisolated from pre-enriched exosome samples using magnetic beads coated with anti-human CD9 or CD81. Exosomes were stained for exosomal antigens CD9 (B and D) and CD81 (C and E) followed flow cytometric analysis and presented as Median Fluorescence Intensity (MFI). A) Scatter plot of exosomes isolated with anti-CD81 and labelled for CD9 to demonstrate gating. Shown is one representative result out of three independent experiments.
Exosomes derived from Jurkat cells were isolated using magnetic beads and prepared for downstream Western blotting analysis. Western blotting requires a larger total surface area for optimal signal. A higher number of magnetic beads were therefore used. In order to illustrate the isolation volume were kept constant in combination with increased amounts of beads. CD81+ exosomes derived from Jurkat cells (Figure 4A) were isolated using magnetic beads coated with anti-human CD81 antibodies at a concentration equal to the concentration used for detection by flow cytometry (1x) in addition to 5x, 10x and 25x the amount of beads used for flow detection. The exosomes were processed for SDS-PAGE and Western blotting and labeled for CD81. The data showed that exosomes isolated with a bead concentration equal to what is used for flow cytometry were not detectable by western blotting. However, increasing amount of CD81 was observed when the amount of beads was increased. Finally, isolation efficiency was confirmed by analysis of the remaining exosomes in the supernatant post isolation measured by flow cytometry (Figure 4B). Increasing amounts of magnetic beads have been used for isolation starting with amounts equivalent to what is used for flow cytometry (1x) up to amount of beads typically used for Western blotting (25x).
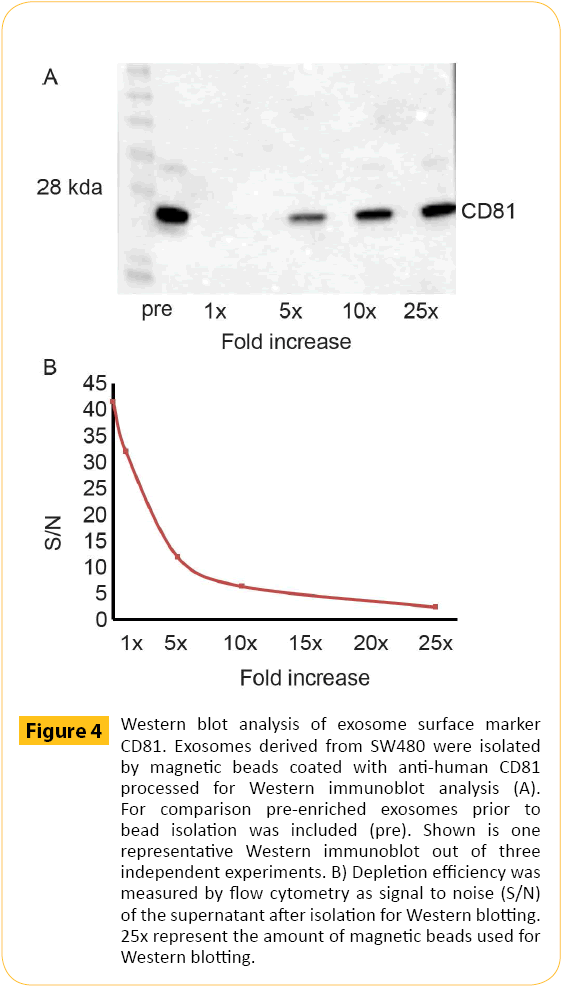
Figure 4: Western blot analysis of exosome surface marker CD81. Exosomes derived from SW480 were isolated by magnetic beads coated with anti-human CD81 processed for Western immunoblot analysis (A). For comparison pre-enriched exosomes prior to bead isolation was included (pre). Shown is one representative Western immunoblot out of three independent experiments. B) Depletion efficiency was measured by flow cytometry as signal to noise (S/N) of the supernatant after isolation for Western blotting. 25x represent the amount of magnetic beads used for Western blotting.
Critical factors for direct isolation of exosomes from cell culture media
In order to provide a fast and scalable protocol compatible with many downstream applications a direct approach would be beneficial. This requires attention to several factors. In order to illustrate this, exosomes derived from SW480 cells were isolated with magnetic beads coated with anti-human CD9 antibodies and subjected to different experimental conditions. First, the impact of volume of exosome containing cell culture media during isolation was addressed. Increased amount of exosome containing cell culture media (175 μl, 350 μl, 700 μl) keeping the amount of magnetic beads constant (20 μl) provided a non-linear (+) dose response curve (Figure 5A). At very high volumes, the reduced concentration of magnetic beads resulted in lower binding kinetics. Secondly, we looked at the effect of increased incubation time. Increased incubation time in combination with fixed amount of exosome containing cell culture media and magnetic beads (20 μl) also gave a non-linear (+) dose response curve (Figure 5B). Finally, we kept the isolation volume and the amount of magnetic bead constant and increased the amount of exosomes in order to ensure equal binding kinetics. The data demonstrated a linear (+) dose response curve (Figure 5C). For sample comparison such experimental settings are critical as it ensures equivalent binding kinetics.
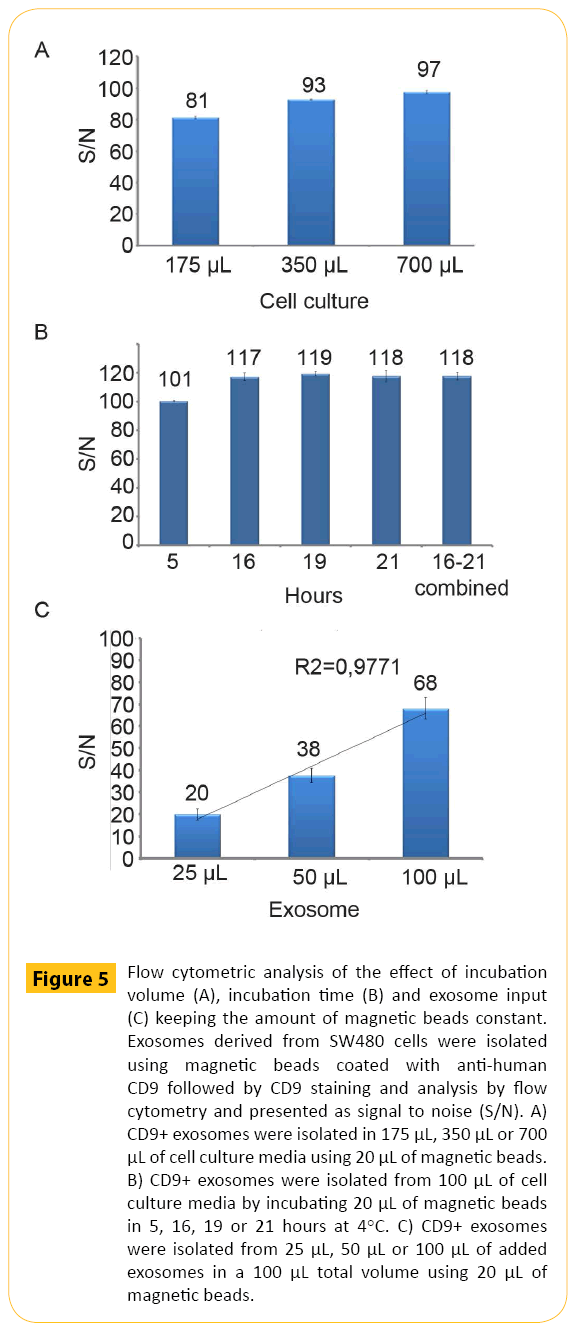
Figure 5: Flow cytometric analysis of the effect of incubation volume (A), incubation time (B) and exosome input (C) keeping the amount of magnetic beads constant. Exosomes derived from SW480 cells were isolated using magnetic beads coated with anti-human CD9 followed by CD9 staining and analysis by flow cytometry and presented as signal to noise (S/N). A) CD9+ exosomes were isolated in 175 μL, 350 μL or 700 μL of cell culture media using 20 μL of magnetic beads. B) CD9+ exosomes were isolated from 100 μL of cell culture media by incubating 20 μL of magnetic beads in 5, 16, 19 or 21 hours at 4°C. C) CD9+ exosomes were isolated from 25 μL, 50 μL or 100 μL of added exosomes in a 100 μL total volume using 20 μL of magnetic beads.
Direct isolation of exosomes from cell culture media
The optimized conditions were used for direct isolation from SW480 cell culture media for flow analysis. SW480 derived exosomes were isolated targeting CD9 followed by CD9 staining. For comparison we related direct (Direct) isolation with exosomes pre-enriched by ultracentrifugation (UC) and precipitation (Prec). To enable direct comparison of the flow analysis the exosome input was normalized for the three ways of processing conditioned cell culture medium based on the concentration factor achieved for UC and prec., respectively.
The results obtained by flow cytometry (Figure 6A) were supported by Western blotting (Figure 6B) demonstrating CD9 staining after direct isolation of CD9+ exosomes (lane 1). Furthermore, CD9 staining was somewhat reduced for UC pre-enriched samples compared to direct isolation indicating loss of exosomes during UC pre-enrichment (lane 2). For isolation from exosomes preenriched by precipitation (lane 3), CD9 staining was comparable to direct isolation (lane 1) . The isolation efficiency was confirmed by flow cytometry analysis. The supernatant after isolation for Western blotting in Figure 6B was subjected to a second round of isolation for downstream flow cytomtery (Figure 6C).
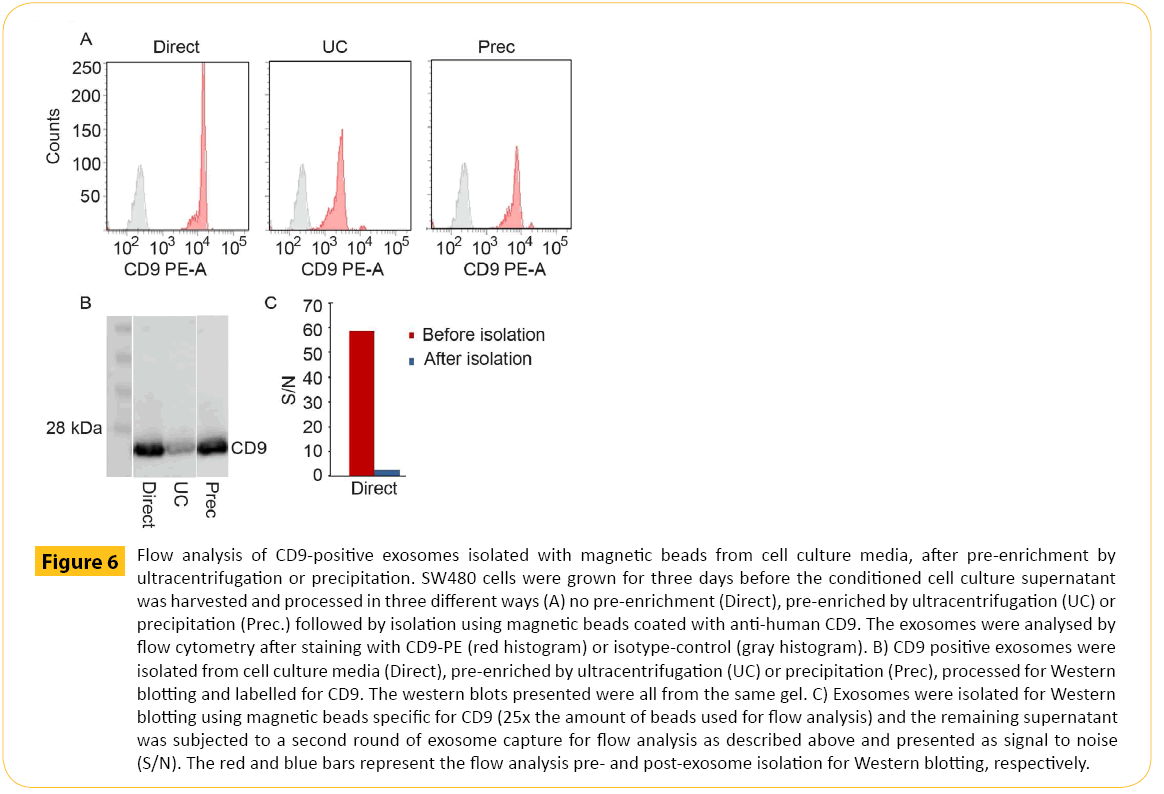
Figure 6: Flow analysis of CD9-positive exosomes isolated with magnetic beads from cell culture media, after pre-enrichment by ultracentrifugation or precipitation. SW480 cells were grown for three days before the conditioned cell culture supernatant was harvested and processed in three different ways (A) no pre-enrichment (Direct), pre-enriched by ultracentrifugation (UC) or precipitation (Prec.) followed by isolation using magnetic beads coated with anti-human CD9. The exosomes were analysed by flow cytometry after staining with CD9-PE (red histogram) or isotype-control (gray histogram). B) CD9 positive exosomes were isolated from cell culture media (Direct), pre-enriched by ultracentrifugation (UC) or precipitation (Prec), processed for Western blotting and labelled for CD9. The western blots presented were all from the same gel. C) Exosomes were isolated for Western blotting using magnetic beads specific for CD9 (25x the amount of beads used for flow analysis) and the remaining supernatant was subjected to a second round of exosome capture for flow analysis as described above and presented as signal to noise (S/N). The red and blue bars represent the flow analysis pre- and post-exosome isolation for Western blotting, respectively.
Finally, CD9+ exosomes isolated with magnetic beads were subjected to immunoassay (Figure 7). First, the correlation between results obtained by flow cytometry and immunoassay were established. CD9+ exosomes stained for CD81 (Figure 7A) and CD9 (Figure 7B) showed high degree of correlation. Secondly, the dose response was investigated. Increasing amount of cell culture media containing exosomes (two separate exosome batches (Batch A and Batch B) were isolated using magnetic beads targeting CD9 followed by CD81 (Figure 7C) or CD9 (Figure 7D) detection. As observed for flow cytometry, a dose response was observed for immunoassay
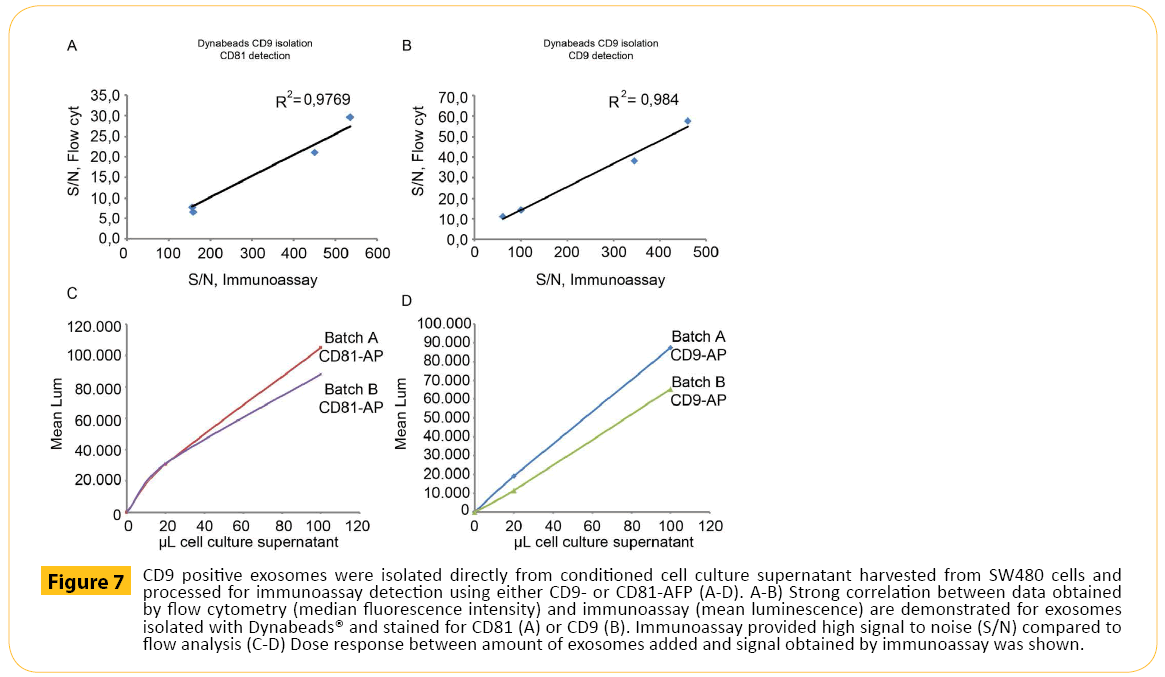
Figure 7: CD9 positive exosomes were isolated directly from conditioned cell culture supernatant harvested from SW480 cells and processed for immunoassay detection using either CD9- or CD81-AFP (A-D). A-B) Strong correlation between data obtained by flow cytometry (median fluorescence intensity) and immunoassay (mean luminescence) are demonstrated for exosomes isolated with Dynabeads® and stained for CD81 (A) or CD9 (B). Immunoassay provided high signal to noise (S/N) compared to flow analysis (C-D) Dose response between amount of exosomes added and signal obtained by immunoassay was shown.
Discussion
Isolation and analysis of nano-sized exosomes is challenging. Today the general approach for exosome isolation is differential centrifugation. The protocol is not standardized as many variants with potential effect on the outcome are used and published. Also, several centrifugation steps are labour intensive, require costly equipment, increase risk for loss of exosomes and does not discriminate well between exosomes and contaminating structures such as larger vesicles and protein/lipid aggregates [26]. The inclusion of magnetic beads with antibodies specific for exosome surface proteins in the isolation protocol downstream of pre-enrichment has successfully overcome the issue of low purity.
Here we provide a simplified exosome isolation workflow by omitting the commonly used pre-enrichment step. We demonstrate that capturing exosomes directly from cell culture supernatant provides sufficient amounts of exosome for downstream analysis while dramatically simplify the sample preparation/handling.
Direct isolation with magnetic beads requires minimal hands-on, provides highly pure exosomes with minimal loss and enables future automation opportunities as it is compatible with the immunoassay format.
Acknowledgements
The authors would like to thank Lisbeth Larsen (Thermo Fisher Scienfic) for technical assistance. B. Kierulf was responsible for the flow assay development and anlysis, flow assay validation flowcyt analysis and data interpretation. A. Kullman was responsible for the western blotting and flow cytometry analysis. M.P. Oksvold was responsible for western blotting and data interpretation. M.Li was responsible for the miRNA analysis. Norbert Roos was responsible for the electronmicroscopy. A.V. Vlassov was responsible for the experimental design and miRNA analysis. Ingrid Manger was responsible for the experimental design of immunoassay and analysis of immunoassay. A.Neurauter was responsible for the experimental design and data interpretation. K.W. Pedersen was the project lead, and responsible for the experimental design, microscopy, data interpretation and writing of the manuscript.
Conflicts of interest
Anette Kullmann, BenteKierulf, Axl Neurauter, Mu Li, Alexander V. Vlassov, Ingrid Manger and Ketil W. Pedersen are full-time employees of Thermo Fisher Scienfic, Oslo, Norway. The authors have indicated that they have no other conflicts of interest regarding the content of this article.
6760
References
- Bobrie A, Colombo M, Raposo G, Thery C (2011) Exosome secretion: molecular mechanisms and roles in immune responses. Traffic12: 1659-1668.
- Thery C, Zitvogel L, Amigorena S (2002) Exosomes: composition, biogenesis and function. Nature reviews. Immunology2: 569-579.
- Janowska-Wieczorek A, Wysoczynski M, Kijowski J, Marquez-Curtis L, Machalinski B, et al. (2005) Microvesicles derived from activated platelets induce metastasis and angiogenesis in lung cancer. International journal of cancer. Journal international du cancer 113: 752-760.
- Belting M, Wittrup A (2008) Nanotubes, exosomes, and nucleic acid-binding peptides provide novel mechanisms of intercellular communication in eukaryotic cells: implications in health and disease. The Journal of cell biology183: 1187-1191.
- Pegtel DM, van de Garde, Middeldorp JM (2011) Viral miRNAs exploiting the endosomal-exosomal pathway for intercellular cross-talk and immune evasion. Biochimica et biophysica acta1809: 715-721.
- Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, et al. (2007) Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature cell biology9: 654-659.
- Al-Nedawi K, Meehan B, Micallef J, Lhotak V, May L, et al. (2008) Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nature cell biology10: 619-624.
- Mittelbrunn M, Gutierrez-Vazquez C, Villarroya-Beltri C, Gonzalez S, Sanchez-Cabo F, et al. (2011) Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nature communications 2: 282.
- Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, et al. (2008) Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nature cell biology10: 1470-1476.
- Zomer A, Vendrig T, Hopmans ES, van Eijndhoven M, Middeldorp JM, et al. (2010) Exosomes: Fit to deliver small RNA. Communicative &integrative biology 3:447-450.
- Lasser C, Eldh M, Lotvall J (2012) Isolation and characterization of RNA-containing exosomes. Journal of visualized experiments: JoVEe3037.
- Conde-Vancells J, Rodriguez-Suarez E, Embade N, Gil D, Matthiesen R, et al. (2008) Characterization and comprehensive proteome profiling of exosomes secreted by hepatocytes. Journal of proteome research7: 5157-5166.
- Subra C, Grand D, Laulagnier K, Stella A, Lambeau G, et al. (2010) Exosomes account for vesicle-mediated transcellular transport of activatable phospholipases and prostaglandins. Journal of lipid research51: 2105-2120.
- Mathivanan S, Simpson RJ (2009) ExoCarta: A compendium of exosomal proteins and RNA. Proteomics9: 4997-5000.
- Thery C, Amigorena S, Raposo G, Clayton A (2006) Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Current protocols in cell biology/editorial board, Juan S. Bonifacinoet alChapter 3, Unit 322.
- Cheruvanky A, Zhou H, Pisitkun T, Kopp JB, Knepper MA, et al.(2007) Rapid isolation of urinary exosomal biomarkers using a nanomembrane ultrafiltration concentrator. American journal of physiology. Renalphysiology 292: F1657-1661.
- Li Q, Eades G, Yao Y, Zhang Y, Zhou Q (2014) Characterization of a stem-like subpopulation in basal-like ductal carcinoma in situ (DCIS) lesions. The Journalof biological chemistry 289: 1303-1312.
- Munoz JL, Bliss SA, Greco SJ, Ramkissoon SH, Ligon KL, et al. (2013) Delivery of Functional Anti-miR-9 by Mesenchymal Stem Cell-derived Exosomes to Glioblastoma Multiforme Cells Conferred Chemosensitivity. Molecular therapy. Nucleic acids2: e126.
- Zeng L, Wang G, Ummarino D, Margariti A, Xu Q, et al. (2013) Histone deacetylase 3 unconventional splicing mediates endothelial-to-mesenchymal transition through transforming growth factor beta2. The Journal ofbiological chemistry 288: 31853-31866.
- Chugh PE, Sin SH, Ozgur S, Henry DH, Menezes P, et al. (2013) Systemically circulating viral and tumor-derived microRNAs in KSHV-associated malignancies. PLoS pathogens9: e1003484.
- Clayton A, Court J, Navabi H, Adams M, Mason MD, et al. (2001) Analysis of antigen presenting cell derived exosomes, based on immuno-magnetic isolation and flow cytometry. Journal of immunologicalmethods 247: 163-174.
- Oksvold MP, Kullmann A, Forfang L, Kierulf B, Li M, et al. (2014) Expression of B-cell surface antigens in subpopulations of exosomes released from B-cell lymphoma cells. Clinical therapeutics 36: 847-862 e841.
- Zeringer E, Li M, Barta T, Schageman J, Pedersen KW, et al. (2013) Methods for the extraction and RNA profiling of exosomes. World journal of methodology3: 11-18.
- Ericsson M, Cudmore S, Shuman S, Condit RC, Griffiths G, et al. (1995) Characterization of ts 16, a temperature-sensitive mutant of vaccinia virus. Journal of virology 69: 7072-7086.
- Yoshioka Y, Konishi Y, Kosaka N, Katsuda T, Kato T, et al. (2013) Comparative marker analysis of extracellular vesicles in different human cancer types. Journal of extracellular vesicles 2.
- Tauro BJ, Greening DW, Mathias RA, Ji H, Mathivanan S, et al. (2012) Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods56: 293-304.












