Keywords
Integron; Dihyrofolate reductase (dfr); Multidrug resistance (MDR); Gene cassette; Enterobacteriaceae
Introduction
Integrons are mobile genetic elements which play a vital role in rapid dissemination of multidrug resistance gene (MDR) among Enterobacteriaceae [1]. The selection processes suggest their accumulation may not involve a single mechanism, but augmented by transposon and plasmids [2]. Integrons carry more than one resistance genes in the form of tandem gene cassettes, mainly based on three essential components in their 5´Conserved Segment (CS): intI gene for encoding enzyme integrase, attI site for site specific recombination and transcription is initiated by a strong promoter Pc upstream of the gene cassette and the 3´CS confers the presence of sulphonamide resistance gene (sul1) and qacEΔ1 genes encoding resistance to quaternary ammonium compounds [3,4]. Based on integrase genes (intI) integrons have been grouped into four classes (Class 1,2,3 and 4) [4] and the genetic arrangement of Class I integrons are strongly associated with multidrug resistance in clinical environment [5]. Acquisition of trimethoprim resistance dihydrofolate reductase (dfr) gene in Enetrobacteriaceae is most other encoded by Integron system [6]. Previously 17 types of trimethoprim resistance dfr genes have been reported [6]. Two more dfr gene was reported recently; found associated with common region (CR1) an IS91- related element in the downstream of 3´CS in integron viz: dfrA24 and dfrA26 [7]. Carriage of other resistant genes as cassette within class I integron, forms a multidrug resistant phenotype, which severely restricts treatment alternatives in clinical settings [7]. So, the current study aim to find out the presence of integron harbouring–positive isolates as well as mapping of whole class 1 integron and to explore the prevalence of integron mediated dfr variants, with special reference with trimethoprimsulphomethoxazole resistant isolates in hospital environment.
Methods
Collection of samples
A total of 268 consecutive non-duplicate clinical isolates of Enterobacteriaceae were taken from the patients admitted to various wards or attended clinics of Silchar medical college and Hospital, Silchar, India from March 2013 to February 2014 (Table 1). Bacterial isolation and identification were according to microscopical investigation, cultural characteristics and biochemical typing [8]. Antibiotic susceptibility was performed against ampicillin (10 μg), co-trimoxazole (1.25/23.75 μg), ciprofloxacin (5 μg), gentamicin (10 μg) and cefepime (30 μg) [Hi- Media, Mumbai, India], by Kirby Bauer disc diffusion method and results were interpreted according to CLSI criteria [9] (Table 2). Escherichia coli ATCC 25922 was taken as negative control.
| Isolates |
Clinical Samples |
| Urine |
Pus |
Stool |
Ear swab |
Throat swab |
Oral swab |
Sputum |
Drain tip |
Blood |
Pseudo pancreatic fluid |
Wound aspirate |
Nasal secretion |
Escherichia.coli
n= 174 |
104 |
40 |
11 |
2 |
6 |
4 |
2 |
1 |
0 |
1 |
2 |
1 |
Klebsiella
Pneumonia
n= 71 |
42 |
18 |
0 |
2 |
4 |
0 |
3 |
1 |
0 |
0 |
0 |
1 |
| Proteus mirabilis n=23 |
3 |
12 |
2 |
1 |
1 |
3 |
0 |
0 |
1 |
0 |
0 |
0 |
Table 1: List of clinical samples showing their antibiotic susceptibility pattern.
| Isolates |
Antibiotics |
| |
Co-trimoxazole |
Ampicillin |
Ciprofloxacin |
Gentamicin |
Cefepime |
Escherichia.coli
n= 174 |
167 |
96% |
142 |
82% |
125 |
72% |
88 |
51% |
83 |
48% |
Klebsiella
Pneumonia
n= 71 |
49 |
69% |
44 |
62% |
35 |
49% |
29 |
41% |
36 |
47% |
| Proteus mirabilis n=23 |
2 |
96% |
21 |
91.3% |
15 |
65% |
15 |
65% |
14 |
61% |
Table 2: Antibiotic susceptibility pattern.
Determination of trimethoprim resistance
All the co-trimoxazole resistant isolates were further tested for susceptibility towards trimethoprim (10 μg) (Hi-Media, Mumbai, India). For further confirmation, MICs of trimethoprim were detected by Hi-Comb MICs Strip (Hi-Media, Mumbai, India).
Screening of class I integron
DNA templates were prepared by using boiling centrifugation method. Overnight culture was suspended in 200 μl sterile distilled water and heated to 80?C for 10 min. Cell suspensions were centrifuged and the supernatant was used for PCR. The presence of class I integron was screened by multiplex PCR using primer int1 and int2 [10] (Table 3). Each single reaction mixture (25 μl) contained 1μl of DNA suspension, 10 pmol of each primer, 2x GoTaq Green Master Mixture [Promega, Madison, USA]. The PCR conditions were as follows; 94°C for 3 minutes, followed by 32 cycles at 94°C for 20 seconds, 54°C for 20 seconds, 72°C for 1 minute and final extension at 72°C for 5 minutes.
| Primer |
Neucleotide Sequence (5′ to 3′) |
Product size (bp) |
|
| Int 1 F |
CAG TGG ACA TAA GCC TGT TC |
160 |
[10] |
| Int 1 R |
CAG TGG ACA TAA GCC TGT TC |
| Int 2 F |
TTG CGA GTA TCC ATA ACC TG |
288 |
[10] |
| Int 2 R |
TTA CCT GCA CTG GAT TAA GC |
| 5-CS |
GGC ATC CAA GCA GCA AG |
[11] |
| 3-CS |
AAG CAG ACT TGA CCT GA |
Table 3: List of Primers.
Mapping of class I integron
PCR was performed with presumptively class I integrase positive isolates using primers 5´– CS GGC ATC CAA GCA GCA AG and 3´– CS AAG CAG ACT TGA CCT GA [11] (Table 3) to amplify the variable region of integron. Each single reaction mixture (30μl) contained 1 μl of DNA suspension, 10 pmol of each primer, 2 x GoTaq Green Master mixtures [Promega, Madison, USA]. The PCR conditions were as follows 95°C for 2 minutes, followed by 35 cycles at 95°C for 20 seconds, 50°C for 45 seconds, 72°C for 2 minute and final extension at 72°C for 7 minutes.
DNA sequence analysis
A number of PCR product that appeared as a unique size of band on the gel was processed with the QIA quick PCR purification Kit (QIAGEN, Germany) and used for direct sequencing. Sequences were compared using BLAST search (https://www.ncbi.nlm.nih.gov/BLAST).
Cloning of class I integron
In order to determine the genetic array of each gene cassettes within the variable region of class I integron amplified products were cloned using pGEM-T vector [Promega, Madison, USA] and transformed into E.coli, JM107. The transformants were subjected to antimicrobial susceptibility testing.
Results
Antibiotic susceptibility pattern
Isolated bacteria (n=268), were identified as Escherichia coli 174 (64.9%), klebsiella pneumoniae 71 (26.5%) and Proteus mirabilis 23 (8.6%) (Table 2). Majority of the bacterial isolates were obtained from urine, followed by pus, stool and other clinical specimens (Table 1). Results of susceptibility profiling showed that, high levels of resistance were recorded towards co-trimoxazole (89%), ampicillin (77%), ciprofloxacin (65%), cefepime (50%) and gentamicin (49%) (Table 2).
PCR screening for integrons
A total of 187 isolates had integron, among which 149 isolates had only class 1 integron, 32 isolates had class 1 integron and class 2 integron, while 11 isolates had only class II integron (Table 4, Figure 1 ).
| Clinical specimen |
Int1 |
Int 2 |
Int 1 and int2 |
| Escherichiacoli= 174 |
98 |
11 |
23 |
| Klebsiella.pneumoniae =71 |
44 |
2 |
4 |
| Proteus mirabilis=23 |
7 |
2 |
5 |
| Total= |
149 |
15 |
32 |
Table 4: Characterization and screening of Integron in Clinical Isolates.
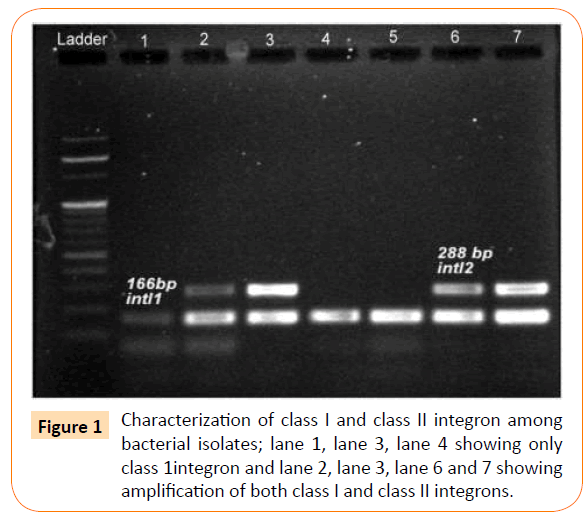
Figure 1: Characterization of class I and class II integron among bacterial isolates; lane 1, lane 3, lane 4 showing only class 1integron and lane 2, lane 3, lane 6 and 7 showing amplification of both class I and class II integrons.
Mapping of class 1 integron
In amplifying the whole class I integron gene cassette for PCR, 95 isolates revealed visible amplification. Variable sizes of class I integrons was between 800 bp-2.5 kb (Figure 2) Sequencing results revealed five different arrays of variable regions and four different types of trimethoprim resistance determinants in gene cassette i.e; dfrA1, dfrA12, dfrA17, dfrA30 (Figure 3). In arrangement I associated with dfrA17 and aadA5 cassette array, and found to be the most prevalent types of gene cassette array (n=44). Whereas, in arrangement II dfrA30, aadA5 in the downstream of the integron I in one isolates. The arrangement III showed the presence of orfF in between of two resistant variants dfrA12 and aadA2 and as well. In arrangement IV, association of dfrA1 and aadA1 gene was observed and this association could be observed, which was observed only in one isolate. The V arrangement showed presence of aacA7 and aadA6 with two chronological 3´-conserved sequences which indicates the association of more than one transposable mechanism.

Figure 2: Amplification of class 1 integron; lane 1, 2, 4 and 6 showing amplification with 1.5 kb and other ranges from 800 bp.
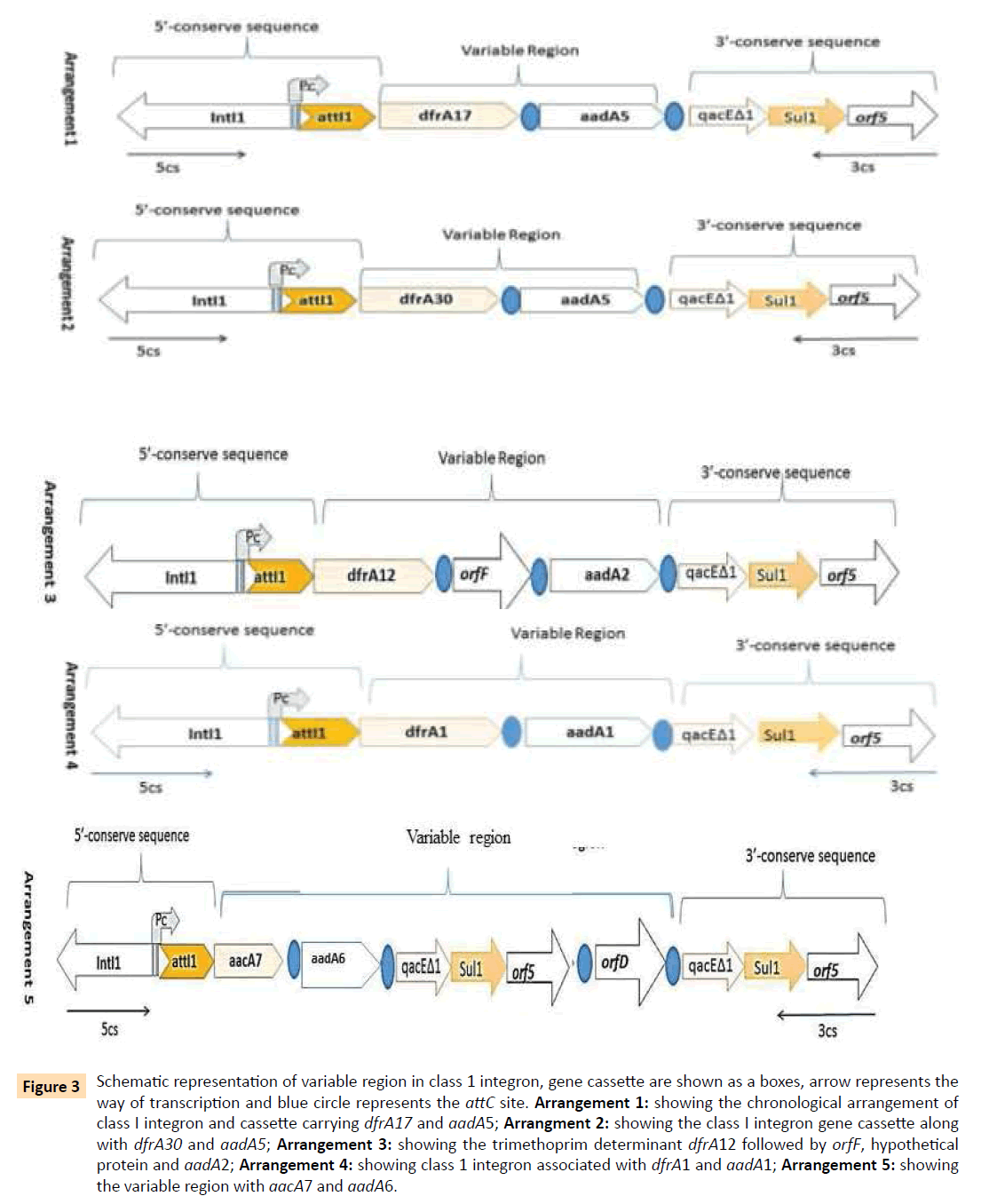
Figure 3: Schematic representation of variable region in class 1 integron, gene cassette are shown as a boxes, arrow represents the way of transcription and blue circle represents the attC site. Arrangement 1: showing the chronological arrangement of class I integron and cassette carrying dfrA17 and aadA5; Arrangment 2: showing the class I integron gene cassette along with dfrA30 and aadA5; Arrangement 3: showing the trimethoprim determinant dfrA12 followed by orfF, hypothetical protein and aadA2; Arrangement 4: showing class 1 integron associated with dfrA1 and aadA1; Arrangement 5: showing the variable region with aacA7 and aadA6.
When tested for susceptibility of each arrangement that was cloned in E.coli DH5α, resistance towards co-trimoxazole was observed but found to be susceptible against aminoglycoside group of antibiotics.
Discussion
This study given a representative structure of the prevalence of integron mediated dfr gene among clinical isolates of Enterobacteriaceae in a hospital setting of Silchar, India. The existence of integron was confirmed in 73.1% of identified multiple antibiotic resistant Enterobacteriaceae strains, which possibly indicates maintenance and propagation of resistance determinants in clinical environment [12,13]. The presence of class I integron were predominant in the Enterobacteriaceae isolates (79.7%). Co-trimoxazole is prescribed for the treatment of urinary tract infection but recently nitrofurantoin, fluoroquinolones and cephalosporins are also being advised, suggesting that intake of these drugs might be sufficient to maintain the antibiotic stress in the clinical settings. Few isolates failed to amplify with 5´-CS and 3´-CS primer, these phenomena could be due to alteration, which occurs in the primer target sites or which are too large to amplify [13]. All the amplification negative isolates were found to confer high level of resistance in one or more antibiotics phenotypically, therefore it could be assumed that the resistance gene might be co-selected with the integrons in another mechanism. It is reported that in rare occasion 3´-CS of class I integron mobilized without any trace of a resistance gene cassette in its surrounding environment [12]. Sequence analysis identified different dfr (dfrA17, dfrA30, dfrA12, dfrA1), four variants of aadA(aadA6, aadA5, aadA2, aadA1) and one aacA7gene (confers resistance to modern aminoglycosides such as amikacin and netilmicin) in gene cassette [13,14]. However, five different arrangements which confer antibiotics resistant to trimethoprim, spectinomycin, streptomycin and one isolates carrying two intact 3´-CS regions; thereby, indicating variability in their origin and selection were recorded in this study. Association of gene cassette with dfrA17 and dfrA12 were the most prevalent in Enterobactericeae and frequently observed to be disseminated through horizontal gene transfer [6]. Gene cassette such as dfrA1- aadA1, dfrA12-orfF-aadA2, dfrA17-aadA5were first observed in 1990 [14,15]. The present study however, showed that the presence of association with integron mediated dfrA30, which was first identified in Klebsiella pneumonia in 2011[15] could not be established. The high correlation between dfrA gene and integron was also reported in different studies [6,14,16,17]. The wide dissemination of dfrA17 variants were found along with aadA5 gene over the course of time [18] and new variants dfrA30 was found associated with integron system. The presence study could highlight parallel existence of five different cassette array systems in a single hospital setting with multiple variants of same resistance gene types (dfr). This kind of study is unique of its own where diverse gene cassettes are maintained in a hospital setting where antibiotic pressure is high.
Conclusion
In conclusion, this study demonstrated the importance of class 1 integrons for acquisition of resistance gene with special reference to trimethoprim, among Enterobacteriaceae. The wide dissemination of dfrA17 variants were found along with aadA5 gene over the course of time and new variants dfrA30 was found associated with integrons system. By using the PCR we have determined the content and order of the antibiotic resistance genes inserted between the conserved regions in the integron system of clinical settings. In the form of gene cassette, selective pressure from one antibiotic can simultaneously select genes when physically linked with other genes. Therefore, PCR mapping with 5´-CS and 3´-CS reveals the position and orientation of inserted gene cassette in integron which can be useful to study the evolution and dissemination of multidrug resistance genes in bacterial population.
Acknowledgement
The authors would like to acknowledge the help of HOD, Microbiology, Assam University for providing infrastructure. The authors sincerely acknowledge the financial support provided by Science and Engineering Research Board (SERB) SR/FT/LS-72/2012 New Delhi, to carry out the work. Authors also acknowledge the help from Assam University Biotech Hub for providing laboratory facility to complete this work.
Conflicts of interest
None to declare
7470
References
- Recchia GD, Hall RM (1995) Gene cassettes: a new class of mobile element. Microbiology 141: 3015-3027.
- Solberg OD, Ajiboye RM, Riley LW (2006) Origin of class 1 and 2 integrons and gene cassettes in a population-based sample of uropathogenic Escherichia coli. J ClinMicrobiol 44: 1347-1351.
- Hall RM, Collis CM (1995) Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. MolMicrobiol 15: 593-600.
- Fluit AC, Schmitz FJ (2004) Resistance integrons and super-integrons. ClinMicrobiol Infect 10: 272-288.
- Martinez FP, Fluit AC, Schmitz FJ, Verhoef J, Jones ME (1999)Many class I integrons comprise distint stable structures occurring in different species of Enterobacteriaceae isolated from widespread geographic regions in Europe. Antimicrob Agents Chemother 43:686-689.
- YU HS, Lee JC, Kang HY, Jeong YS, Lee EY, et al. (2004) Prevalence of dfr Genes Associated with Integrons and Dissemination of dfrA17 Among Urinary Isolates of Escherichia coli in Korea. J AntimicrobChemother53:445-450.
- Grape M, Sundström L, Kronvall G (2007) Two new dfr genes in trimethoprim-resistant integron-negative Escherichia coli isolates. Antimicrob Agents Chemother 51: 1863-1864.
- Clinical and laboratory Standard Institute (2011). Performance Standards for Antimicrobial Susceptibility Testing: Twenty –first Informational Supplement. CLSI, Wayne, P.A USA: M100-S21.
- Colee JG, Diguid JP, Fraser AG (1996). Mackie and McCartney Practical Medical Microbiology 14thedn, Edinburgh, Churchill, Livingstone.
- Koeleman JG, Stoof J, Van Der Bijl MW, Vandenbroucke-Grauls CM, Savelkoul PH (2001) Identification of epidemic strains of Acinetobacterbaumannii by integrase gene PCR. J ClinMicrobiol 39: 8-13.
- Levesque C1, Piché L, Larose C, Roy PH (1995) PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob Agents Chemother 39: 185-191.
- Grape M, Farra A, Kronvall G, Sundström L (2005) Integrons and gene cassettes in clinical isolates of co-trimoxazole-resistant Gram-negative bacteria. ClinMicrobiol Infect 11: 185-192.
- White PA, McIver CJ, Deng Y, Rawlinson WD (2000) Characterisation of two new gene cassettes, aadA5 and dfrA17. FEMS MicrobiolLett 182: 265-269.
- White PA, McIver CJ, Rawlinson WD (2001) Integrons and gene cassettes in the enterobacteriaceae. Antimicrob Agents Chemother 45: 2658-2661.
- Kumar A, Chakraborti S, Joshi P, Chakrabarti P and Chakraborty R (2011).A Multiple Antibiotic and Serum Resistant Oligotrophic Strain, klebsiella pneumonia MB45 having novel dfrA30, is sentive to ZnO QDs. Annals AntimicrobChemother: 10-19.
- El-Najjar NG, Farah MJ, Hashwa FA, Tokajian ST (2010) Antibiotic resistance patterns and sequencing of class I integron from uropathogenic Escherichia coli in Lebanon. LettApplMicrobiol 51: 456-461.
- Blahna MT, Zalewski CA, Reuer J, Kahlmeter G, Foxman B, Marrs CF (2006). The role of horizontal gene transfer in the spread of trimethoprim-sulfamethoxazole resistance among uropathogenic Escherichia coli in Europe andCabada. J.AntimicrobChemother57:666-672.
- Adel El-Sokkary MM, Adbelmegged ES (2015). Chracterisation of Class 1 integron among Escherichia coli isolated from Mansoura University Hospitals in Egypt. Adv In Microbio 5:269-277.








