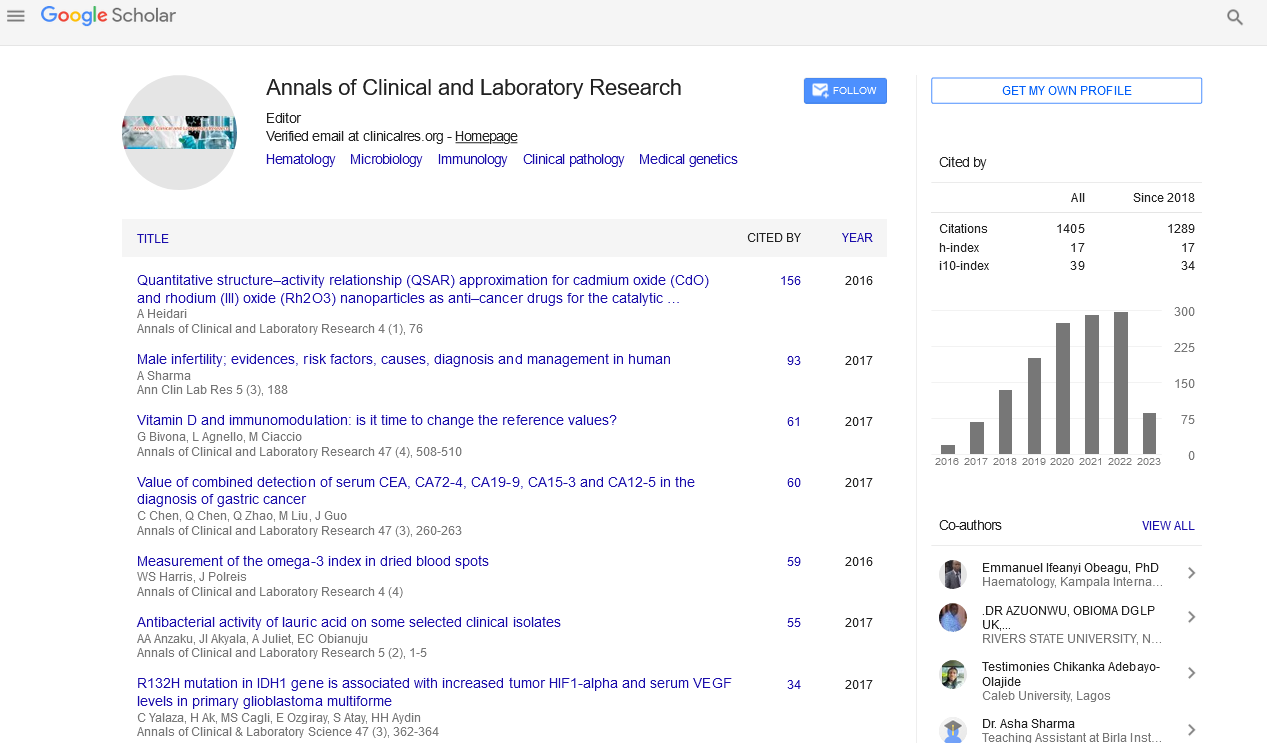
Opinion - (2023) Volume 11, Issue 5
Environmental Epigenetics: Unraveling the Impact of Exposures on Gene Regulation
Menglin Cheng*
Menglin Cheng, Nutrition and Sport, La Trobe University, Bundoora, Australia
*Correspondence:
Menglin Cheng, Menglin Cheng, Nutrition and Sport, La Trobe University,
Australia,
Email:
Received: 09-Sep-2023
Published:
30-Oct-2023, DOI: 10.36648/2386-5180.23.11.488
Introduction
Environmental epigenetics is a burgeoning field that investigates
how environmental factors, from chemicals and pollutants to diet
and lifestyle, can influence gene expression through epigenetic
modifications. Epigenetics refers to changes in gene activities
that do not alter the DNA sequence but rather affect how
genes are turned on or off. These epigenetic changes can have
a profound impact on an individual's health, development, and
susceptibility to diseases. This article delves into the fascinating
realm of environmental epigenetics, exploring how exposures
shape our epigenome and, in turn, our biology [1].
The epigenome serves as a molecular switchboard that controls
gene expression. Two primary mechanisms govern epigenetic
regulation: DNA methylation and histone modifications. This
process involves the addition of methyl groups to specific
regions of DNA, typically cytosine residues in CpG dinucleotides.
Methylation can silence gene expression by preventing the binding
of transcription factors and other regulatory proteins to the gene's
promoter region. Histones are proteins around which DNA is
wound [2]. Chemical modifications of histones, such as acetylation,
methylation, and phosphorylation, can alter the structure of
chromatin, making genes more or less accessible for transcription.
Environmental exposures can induce changes in the epigenome,
leading to altered gene expression profiles. Diet plays a crucial
role in epigenetic regulation. Nutrients like folate, choline, and
methyl donors influence DNA methylation patterns. Conversely,
a diet high in processed foods and low in essential nutrients can
lead to epigenetic changes associated with chronic diseases.
Environmental toxins, such as heavy metals, pesticides, and air
pollutants, can induce epigenetic modifications. For example,
exposure to lead has been linked to DNA methylation changes
that impact cognitive development in children. Chronic stress
and adverse life events can trigger epigenetic changes in genes
associated with the stress response and mental health. These
changes may contribute to conditions like depression and
anxiety. Smoking, alcohol consumption, and substance abuse can
induce epigenetic alterations that increase the risk of cancer and
other diseases [3].
Gut bacteria can produce metabolites that influence epigenetic
regulation in the host. This connection between the micro
biome and epigenetics has implications for metabolic disorders and autoimmune diseases. Understanding the mechanisms by
which environmental exposures influence the epigenome is
a complex and evolving area of research. Some key points to
consider include: Research has identified specific epigenetic
marks associated with various exposures. For instance, exposure
to certain pollutants is linked to altered DNA methylation
patterns in genes involved in inflammation and oxidative stress
[4]. The timing of exposures is critical. Epigenetic changes can
occur during critical periods of development, affecting health
outcomes later in life. For example, maternal smoking during
pregnancy can lead to epigenetic changes in the fetal epigenome,
influencing lung function and increasing the risk of asthma in
the child. Some epigenetic changes induced by environmental
exposures can be passed down to future generations. This trans
generational epigenetic inheritance raises ethical and healthrelated
questions.
Epigenetic modifications can serve as early indicators of disease
risk or environmental exposure. Identifying biomarkers allows for
targeted interventions and personalized medicine approaches.
Understanding how the epigenome responds to environmental
factors opens the door to interventions aimed at reversing or
mitigating epigenetic changes associated with disease. Diet
modifications, lifestyle changes, and even pharmaceutical
interventions may be explored. Research in this field can inform
public policy decisions related to environmental regulations,
nutrition, and public health initiatives aimed at reducing
exposures and mitigating epigenetic risks [5].
Conclusion
Environmental epigenetics is a captivating field that sheds light
on how the environment interacts with our genes to influence
health and disease. By unraveling the impact of exposures on
the epigenome, researchers and healthcare professionals can
better understand the underlying mechanisms of diseases,
develop targeted interventions, and ultimately improve public
health outcomes. As this field continues to advance, it holds the
promise of revolutionizing our approach to disease prevention
and personalized medicine.
References
- Garcia-Fortes M, Hernandez-Boluda JC, Alvarez-Larran A, Raya JM, Angona A, et al (2022). Impact of individual comorbidities on survival of patients with myelofibrosis. Cancers. 14(9):2331.
Indexed at, Google Scholar, Cross Ref
- Masyuko S, Mukui I, Njathi O, Kimani M, Oluoch P, et al (2018). Pre-exposure prophylaxis rollout in a national public sector program: the Kenyan case study. Sexual Health. 15(6):578-86.
Indexed at, Google Scholar, Cross Ref
- Blackledge NP, Klose RJ (2021.) The molecular principles of gene regulation by Polycomb repressive complexes. Nat Rev Mol Cell Biol. 22(12):815-33.
Indexed at, Google Scholar, Cross Ref
- Mir M, Bickmore W, Furlong EE, Narlikar G (2019). Chromatin topology, condensates and gene regulation: shifting paradigms or just a phase?. Development. 146(19):182766.
Indexed at, Google Scholar, Cross Ref
- He L, Hannon GJ (2004). MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 5(7):522-31.
Indexed at, Google Scholar, Cross Ref