Sujith Sugathan1, Aseer Manilal2*, Tigist Gezmu2, Behailu Merdekios2, Joseph Selvin3, Tsegaye Tsalla2, Akbar Idhayadhulla4and Shine Kadaikunnan5
1Division of Microbiology, Department of Botany and Biotechnology, Sree Narayana College, Kollam 691021 Kerala, India
2College of Medicine and Health Sciences, Arba Minch University, Arba Minch, Ethiopia
3Centre for Microbiology, Pondicherry University, Puducherry 605014, South India
4School of Chemical Engineering, Yeungnam University, Gyeongsan 712- 749, South Korea
5Department of Botany and Microbiology, College of Science, King Saud University, P.O. Box 24, Kingdom of Saudi Arabia
*Corresponding Author:
Aseer Manilal
Department of Medical Laboratory Science, College of Medicine and Health Sciences, Arba Minch University, Arba Minch, Ethiopia
Tel: +251-919904201
E-mail: aseermanilal@gmail.com
Received Date: January 24, 2015; Accepted Date: February 10, 2015; Published Date: February 13, 2015
Citation: Sugathan S, Manilal A, Gezmu T, et al. Evaluating the Antibacterial Potential of Streptomyces sp. Transl Biomed. 2015, 6:1. DOI: 10.21767/2172-0479.100003
Keywords
Marine sponge isolates; Streptomyces; Secondary metabolites; Antagonistic activity
Introduction
The available statistics indicate marine invertebrates still to be virgin reservoir with rich novel biomedical leads [1]. Nevertheless, it’s difficult to procure adequate and reliable supplies of these compounds from the nature. Hence, to resolve this tangle, new research focusing on marine microorganisms has been gaining considerable attention [2]. The majority of these organisms have been well known to produce bioactives which assist them in the colonization of surfaces including those of their host animals. During the last few decades probing for therapeutically active substances has riveted on marine organisms as it is relatively easy to isolate, purify and produce in bulk-scale [1]. In contrast with terrestrial organisms, marine organisms produce a cornucopia of bioactive secondary metabolites [3]. The secondary metabolites by marine bacteria isolated from different seas have been reported since long [3]. The majority of these bacterial strains were isolated from seawater and marine sediments. Bacteria associated with marine invertebrates have been found to display signi?cant bioactivity including antifouling, antibacterial and cytotoxic activities. And there are innumerable studies on marine medical exploration and ecology investigation about sponge associated bacteria and fungi [4-7]. Investigation of marine bacteria isolated from marine algae and sponge has revealed many of them having the ability to preclude the growth of human and shrimp pathogens [4,8]. In the light of this, the present research is focused on the screening of antagonistic bacteria isolated from marine sponge against human and shrimp pathogens.
Methods
Collection of sponges
The sponge specimens were collected off the Vizhinjam littoral (80°21’N, 77°00’ E) by snorkeling at a depth of between 1 to 3 m. Upon collection, the sponges were soaked in aged natural sea water for 2 h to remove slackly associated microorganisms [4]. Afterwards, the drained samples were kept in a sterile incubator oven for 1 h at 40°C to dry the surface, and frozen at 20°C.
Isolation of the marine sponge-associated microbes
A dollop of sponge tissue was excised using a sterile scalpel from the internal mesohyl region. The tissue samples were then homogenized with sterile sea water in a tissue homogenizer. Dilution series were prepared from the homogenates and each dilution (~500 μL) was inoculated by spread plate on to the different media such as Zobell marine agar (ZMA) or nutrient agar (1.5% NaCl). These media were supplemented with Nalidixic acid and cyclo hexamide to inhibit the other bacterial and fungal contamination, respectively. The inoculated Petri-plates were incubated for 14 days at 27°C after which colony-forming units (CFU) were counted. Counts between 30 and 200 CFUs were used for analysis. On the basis of morphological features, different colony types were randomly picked and purified by making streak plates on NA plates supplemented with 1.5% of NaCl. The diversity of colonies was examined based on morphological characteristics such as size, color, opacity, texture, form, elevation, margin and surface [9]. Single colonies were transferred at least three times until considered pure. Strains were stored frozen at 4°C in Zobell Marine Broth.
The potential antagonistic bacteria were selected by screening the sponge isolates against a battery of human and shrimp pathogens (Table 1). The human and shrimp pathogens with MTCC number were obtained from the Institute of Microbial Technology (IMTECH), Chandigarh, India. While the Vibrio isolates were culled from Penaeus monodon smote with Vibriosis [10]. All the test organisms were maintained on nutrient agar slants at 4°C before being used in the inhibitory assays. The antimicrobial activities of the sponge associated bacteria were performed by cross-streak method [11]. Mueller-Hinton agar plates were prepared and inoculated with sponge isolates by a single perpendicular streak of inocula in the centre of the Petri-dish and incubated at 27°C for 4 days. The plates were seeded with test organisms by a perpendicular streak at a 90° angle to the line of the sponge isolates. Antagonism was observed based on the inhibitory interaction between the sponge isolates and test organisms. The reference standard (Nalidixic acid) was used for the detection of strain sensitivity/resistance. Inhibition activities and colonization effect are noticed at 24, 48, 72 and 96 hours. All the tests were conducted in triplicates.
| Group |
Species |
Human pathogens
(MTCC) |
Staphylococcus aureus (MTCC 96) |
| Bacillus amyloliquefaciens (10441) |
| Shigellaflexneri(MTCC 1457) |
| Micrococcus luteus(MTCC106) |
| Vibrio mimicus (MTCC 4434) |
| Shrimp pathogens (MTCC) |
V. alginolyticus (MTCC4439) |
| V. vulnificus (MTCC 1145) |
| V. parahaemolyticus(MTCC451) |
| V. alcaligenes(MTCC 4442) |
| V. fischeri(MTCC 1738) |
| V. harveyi(MTCC 3438) |
| V. anguillarum(VB10) |
| Shrimp vibrio isolates |
V. alginolyticus(VB11) |
| V. vulnificus(VB14) |
| V. parahaemolyticus(VB12) |
| V. harveyi(VB15) |
| V. fischeri(VB17) |
| V. splendidus(VB22) |
| V. campbellii(VB23) |
| Ph. damselae(VB26) |
Table 1: Panel of pathogens used for antimicrobial assay.
Antimicrobial screening of sponge-associates by agar well diffusion method
The bacterial strains which displaying the antagonistic activity in cross-streak method was further confirmed with the agar welldiffusion method as described by Manilal et al. [4]. The sponge bacterial strains were grown in a nutrient broth (Merck, Germany), incubated at 25°C for 18h, and adjusted to an approximate concentration of 108 CFU ml–1. The Mueller Hinton agar plates were prepared and uniformly swabbed with respective pathogens. Thereafter, it was punched with a five millimeters diameter wells and filled with 120 μl of the respective microbial cell suspension (CS). The well with broth used for culture was considered as negative control. The Petri-dishes were incubated at 37°C for 24h. After incubation, plates were examined for inhibition zones. The assay was performed in triplicates of individual Petri-dishes. Clear zone of inhibition formed around wells were considered as indicative of antimicrobial activity. The inhibitory activity was measured by calculating the area of clear zone. The antibiogram was statistically analyzed for the determination of Skewness among the tested bacterial strains.
Biochemical profiling of sponge isolates
Biochemical characterization of the potential sponge isolates were performed using the methods described by Lalucat et al. [12] and Lechevalier [13] respectively. Morphological characterization of the sponge isolates were determined by motility test, Gram’s staining and Ziehl Nielsen staining. Physiological characterization was performed by checking the hydrolytic activity and carbohydrate fermentation of the potential antagonistic strains. The cultural characteristics of the sponge isolates were determined by naked eye examination of 14 days old cultures that were grown on different media such as Actinomyces agar, Actinomycetes isolation agar (AI agar), Yeast Malt Extract agar (YME agar), Inorganic salt agar International Streptomyces Project Agar (ISP-4agar), Oat meal agar, Oat meal with glycerol medium and Chitin agar. The micro-morphology and sporulation were observed by light microscopy. Antibiotic sensitivity was also examined using selected antibiotics. Colours of aerial and substrate mycelium were determined with the ISCCNBS centroid colour charts (US National Bureau of Standard, 1976). Hemolytic activity was examined using blood agar plates with 5% human blood. Plates were analyzed for hemolysis after incubation at 37°C for 24 h. The clear zone of clearance around the colony was considered as positive. All the culture media were prepared and treated as per the manufacturer guidelines.
Molecular identification of the active sponge isolate (MAPS 15)
Bacteria with highest and broadest rank of antimicrobial activity and non-hemolytic property was chosen for partial sequencing and phylogenetic analysis. Molecular identification of the potential candidate sponge isolate was based on partial 16S ribosomal RNA gene sequencing. The MAPS 15 bacterial strain was cultured in marine broth at 25°C, and genomic DNA was extracted by the CTAB/ NaCl method. The universal eubacterial 16S rRNA gene primer (5’-3’; 5-3’) was used for the PCR amplification of bacterial 16S ribosomal RNA gene. The amplified 16S ribosomal RNA gene product was cloned by the TA cloning method using a TOPO TA cloning kit as per the manufactures instruction (Invitrogen) for sequencing. The obtained 16S ribosomal RNA gene sequence from the active isolate MAPS 15 was compared with sequences of other bacteria by using BLAST (Basic Local Alignment Search Tool) [14] to analyse pair wise homology. The sequences obtained were deposited in GenBank, EMBL in Europe and the DNA Data Bank of Japan with an accession number JQ723478.
Statistical analysis
All the data were expressed as mean ± standard deviation (S.D.). Mean values were assessed using one way analysis of variance (ANOVA)using SPSS for Windows version 20.0 (Statistical Package for Social Services, Chicago, IL, USA).
Results
Based on the colony morphology observed and stability in subculturing, a total of 51 strains of bacteria were segregated from five different species of marine sponges and designated as MAPS (Marine Actinomycetes Probiotic Strain). The highest number of bacterial strains was isolated from D. nigra (18.6%), followed by C. diffusa (10.6%), F. cavernosa, (8.6%), S. officinalis (8.2%), and Drysidea sp. (6.9%) (Figure 1 and Table 2). Among the media used, ZoBell marine agar (ZMA) was found to be the most appropriate medium for the isolation of sponge-associated bacteria. Sixteen strains of bacteria were isolated on ZMA.
| Marine sponges |
Strains |
| Dendrilla nigra |
9 (MAPS1 to MAPS9) |
| Spongia officinalis |
14 ( MAPS10 to MAPS23) |
| Fasciospongia cavernosa |
10 (MAPS24 to MAPS33) |
| Callyspongia diffusa |
11 (MAPS34 to MAPS44) |
| Drysidea sp. |
7 (MAPS45 to 51) |
*MAPS-Marine actinomycetes probiotic strain.
Table 2: Bacterial strains isolated from marine sponges.
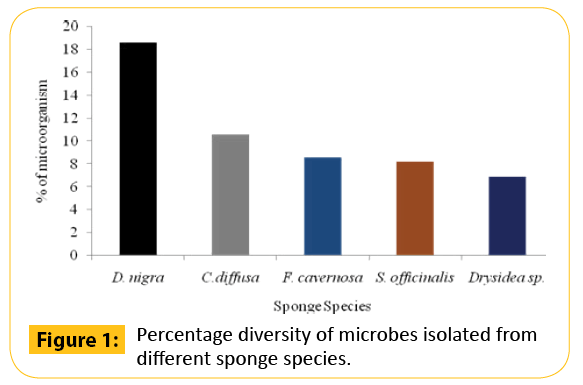
Figure 1: Percentage diversity of microbes isolated from different sponge species.
Antagonistic activity of the marine sponge isolates
In the preliminary antibacterial screening, it was observed that among the fifty one bacteria, only twelve strains of bacteria isolated from marine sponges, S. officinalis and D. nigra exhibited activity against at least one of the tested pathogens while other strains produced no results at all. A higher percentage of antagonistic strains were isolated from S. officinalis. The activity was meager for the rest of the strains isolated from D. nigra. On the basis of results from the cross streak method, only two isolates (MAPS 08 and MAPS 15) were further subjected to agar-well diffusion assay against a battery of human and animal pathogens. Both isolates surpassed the growth of 20 tested bacterial pathogens including type culture of human and shrimp pathogens and vibrio isolates. The anti biogram of MAPS 08 and MAPS 15 revealed that the cell suspension contained a broad-spectrum bactericidal agent. The antibacterial potentials of MAPS 08 and MAPS 15 are depictured in the Figures 2-4. Of the two isolates, MAPS 15 showed the highest activity. The CS of MAPS 15 displayed greater inhibitory effects against the growth of human and shrimp pathogens. The striking inhibition values were recorded against shrimp pathogens such as V. alginolyticus (519.98 ± 23.2 mm2 ) and V. alcaligenes (510.98 ± 22.2 mm2 ). The antibacterial potential of MAPS 15 against the human pathogens was in the range of 156.2 ± 14.4 to 270.6 ± 11.8 mm2 . The highest inhibitory zone of 270.6 ± 11.8 mm2 was manifested against S. aureus whereas the least inhibitory zone was produced against S. flexneri (156.2 ± 14.4 mm2 ). Similarly, the CS of MAPS 08 produced an activity ranged between 132.7 ± 7.3 and 212.9 ± 12.3 mm2 against the human pathogens and 55.8 ± 5.9 to 234.08 ± 10.3 mm2 of inhibitory zone against the shrimp Vibrio isolates and 83.3±5.8 to 473.86 ± 17.3 mm2 for shrimp MTCC cultures. Based on the highest and broadest activity spectrum, MAPS-15 was chosen for further studies.
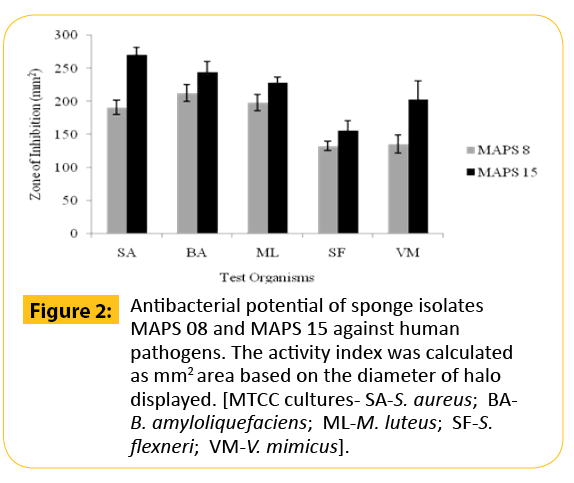
Figure 2: Antibacterial potential of sponge isolates MAPS 08 and MAPS 15 against human pathogens. The activity index was calculated as mm2 area based on the diameter of halo displayed. [MTCC cultures- SA-S. aureus; BAB. amyloliquefaciens; ML-M. luteus; SF-S. flexneri; VM-V. mimicus].
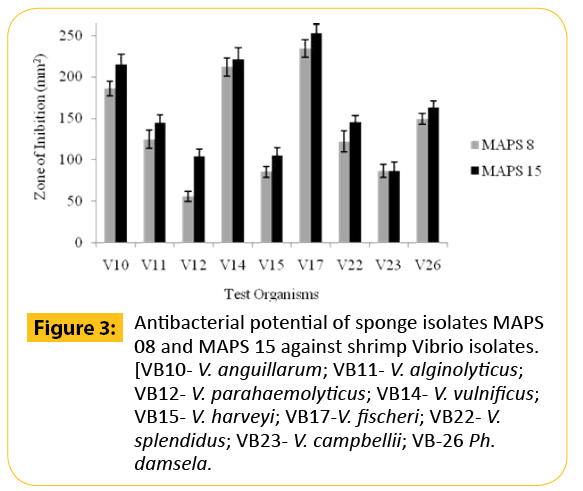
Figure 3: Antibacterial potential of sponge isolates MAPS 08 and MAPS 15 against shrimp Vibrio isolates. [VB10- V. anguillarum; VB11- V. alginolyticus; VB12- V. parahaemolyticus; VB14- V. vulnificus; VB15- V. harveyi; VB17-V. fischeri; VB22- V. splendidus; VB23- V. campbellii; VB-26 Ph. damsela.
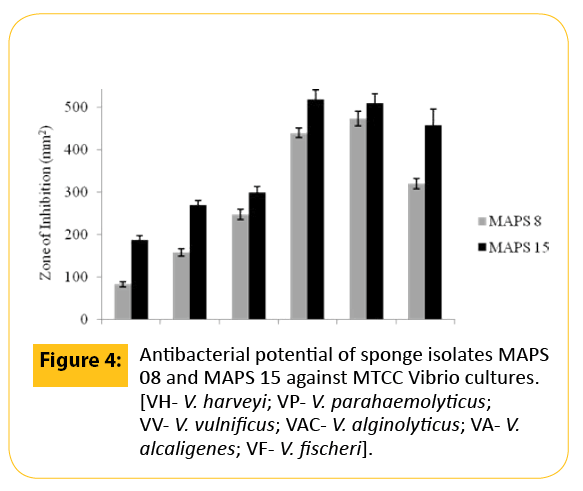
Figure 4: Antibacterial potential of sponge isolates MAPS 08 and MAPS 15 against MTCC Vibrio cultures. [VH- V. harveyi; VP- V. parahaemolyticus; VV- V. vulnificus; VAC- V. alginolyticus; VA- V. alcaligenes; VF- V. fischeri].
Bio physiochemical profiling of the spongeassociated antagonistic bacteria
The morphological, physiological and biochemical profile of the sponge isolates (MAPS 08 and MAPS 15) are presented in Tables 3-6. The results of the morphological characteristics showed that both the strains were non motile gram positive rod shaped microorganisms. The elaborated cultural characteristics of the antagonistic strains were shown in Table 7. Several media such as Zobell Marine Agar, Actinomyces agar, Actinomycete isolation agar, Yeast malt extract agar, Inorganic salts starch medium, Oat meal agar, Oat meal with glycerol medium and chitin agar were used to determine the cultural characteristics of the isolates. The MAPS 08 strain showed maximal growth in all the media used whereas it showed moderate growth in yeast malt extract agar, inorganic salt starch media and chitin agar.
| Strains |
Motility |
Grams staining |
Ziehl Nielsen staining |
| MAPS08 |
*NM |
**G +ve rod |
Aerial mycelium with two branches, conidia present |
| MAPS15 |
NM |
G +ve rod |
Mycelium with branches, conidia present |
Table 3 :Morphological characterization of sponge isolates (MAPS 08 and MAPS 15)
| Strain |
Starch |
Gelatin |
Cellulose |
Tributyrin |
Chitin |
| MAPS 08 |
+ |
+ |
+ |
+ |
+ |
| MAPS 15 |
+ |
+ |
+ |
+ |
- |
Table 4: Physiological characteristics (Hydrolytic activity) of antagonistic strains (MAPS 08 and MAPS 15).
| Strains |
Glucose |
Mannitol |
Sucrose |
Gelatin |
Glycerol |
| MAPS 08 |
+ |
- |
- |
- |
+ |
| MAPS 15 |
+ |
- |
- |
- |
+ |
Table 5: Carbohydrate fermentation of antagonistic strains (MAPS 08 and MAPS 15).
| Strains |
Citrate |
TSI |
MR |
VP |
Oxidase |
Catalase |
Indole |
Hemolyticactivity |
| MAPS08 |
+ |
+ |
+ |
- |
+(Weak) |
- |
- |
+ |
| MAPS15 |
+ |
+ |
+ |
- |
+ |
- |
- |
- |
TSI– Triple Sugar Iron agar test; MR– Methyl Red; VP– Voges Proskauer, +-Positive, – -Negative.
Table 6: Biochemical characteristics of antagonistic strains (MAPS 08 and MAPS 15).
| Strains |
Actinomyces agar; |
Actinomycetes isolation agar |
*YME agar |
**ISP–agar |
Oatmeal agar |
Oat meal with glycerol medium |
Chitin agar |
| MAPS08 |
Aerial pale yellow, large mycelium surrounded, yellowish brown centered powdery colonies. |
Aerial white and substrate yellow powdery colonies. |
Aerial and substrate muddy white color pin headed colonies. |
Pale yellow partially slimy and dry colonies. |
Ash color colonies, less growth. |
Yellow color mucoid flat colonies. |
Ash with yellow slimy colonies. |
| MAPS15 |
Aerial and substrate ash with slimy colonies. |
Aerial white and substrate yellow powdery colonies. |
Observed less growth pale white color powdery colonies. |
Ash with white color minute pin headed dry colonies. |
Ash with yellow color mucoid colonies, less growth. |
Dark yellow Color mucoid, flat colonies |
No growth. |
*YME-Yeast Malt Extract, **ISP-4-Inorganic salt agar International Streptomyces Project Agar (ISP).
Table 7: Cultural characteristics of antagonistic strains (MAPS 08 and MAPS 15).
The strain MAPS 08 appeared as pale yellow colored complex aerial mycelia with white colored mass of spores. The strain showed positive results for starch, gelatin, cellulose, tributyrin and chitin hydrolysis. It was positive for glucose and glycerol fermentation and was negative for mannitol, sucrose and gelatin. The isolates are able to produce enzyme oxidase, but unable to produce positive result for catalase. The MAPS 08 strain was found to be positive for citrate, TSI and MR. It was negative for VP and indole. The antibiotic sensitivity pattern of the isolate is shown in Table 8. The strain was sensitive to ciprofloxacin and chloramphenicol. It showed prominent resistance against ampicillin and tetracycline. A distinct characteristic of MAPS 08 was its ability to produce haemolysin. It exhibited significant hemolytic activity. Therefore, this strain is excluded from further molecular identification. The strain MAPS 15 showed maximal growth in all the media used whereas it showed moderate growth in inorganic salt starch media, less growth in yeast malt extract agar and showed no growth in chitin agar. The strain MAPS 15 appeared as ash colored slimy colony with complex aerial mycelia. The strain showed positive results for starch, gelatin, cellulose, tributyrin but showed negative result for chitin hydrolysis. It was positive for glucose and glycerol fermentation and was negative for mannitol, sucrose and gelatin. The isolates are able to produce enzyme oxidase, but are unable to produce positive result for catalase. The MAPS 15 strain was found to be positive for citrate, TSI and MR. It was negative for VP and indole test. The antibiotic sensitivity pattern of MAPS 15 is shown in Table 8. The strain was sensitive to tetracycline, ciprofloxacin and chloramphenicol. It showed prominent resistance against ampicillin. In hemolysis assay, MAPS 15 showed a negative result.
| Strain |
Ampicillin |
Tetracycline |
Ciprofloxacin |
Chloramphenicol |
| MAPS08 |
*R |
R |
S |
S |
| MAPS15 |
R |
**S |
S |
S |
*R– Resistant; **S-Sensitive
Table 8: Antibiotic sensitivity of the antagonistic strains (MAPS 08 and MAPS 15).
Molecular identifications of the sponge isolate MAPS 15
The genomic DNA isolated and subjected to 16S rRNA amplification for the identification of the potent antagonistic producer. A PCR product of length 1200 bp was purified using the GeneiPureTM quick PCR purification kit and sequenced by Sigma Aldrich. Taxonomic affiliation of the 16S rRNA sequences of the isolate MAPS 15 was retrieved from classifier program of Ribosomal Database Project II release 10.0 (https://rdp.cme.msu.edu/). The 16S rRNA sequence of the isolate was blasted using megablast tool of GenBank (https://www.ncbi.nlm.nih.gov/). This revealed that the isolate was a Streptomyces strain. Representative of maximum homologous (98-99%) sequences of each isolate were obtained from seqmatch programme of RDPII and were used for the construction of phylogenetic affiliation (Figure 5). The 16S rRNA sequence of the isolate MAPS 15 was further analyzed using NCBI BLASTn tool with a query to limit the search to the closest relatives such as Streptomyces erythrogriseus LMG 19406 SCF18 and Streptomyces vinaceus NBRC 3406. Representatives of maximum homologous sequences from the search were used for the construction of phylogenetic tree using UPGMA algorithm. The 16S rRNA gene partial sequence obtained in this study was submitted to the GenBank database with assigned accession numbers JQ723478.
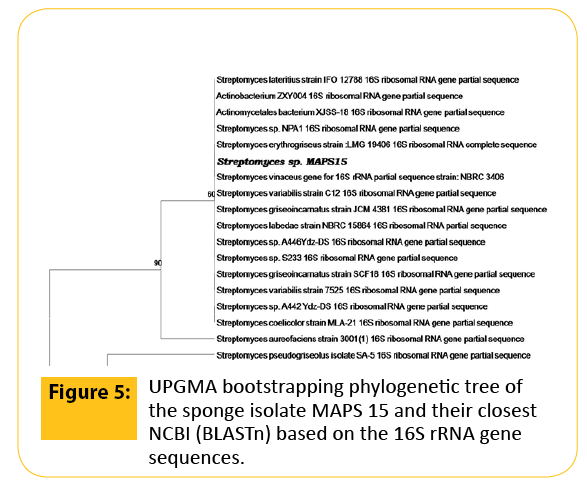
Figure 5: UPGMA bootstrapping phylogenetic tree of the sponge isolate MAPS 15 and their closest NCBI (BLASTn) based on the 16S rRNA gene sequences.
Discussion
It is a long-familiar concept that sponges harbour verdant types of microbial communities with versatile biopotency. Recent years have witnessed the origination and development of novel bio-engineering techniques on the production of bioactive metabolites from marine sponge associated microbes [3]. The studies on bioactivity screening of microorganisms such as this provide a base line data for the identification of possible antagonistic candidates. In the present study, antagonistic effects of sponge isolates on the growth of human and shrimp pathogens were determined. The highest proportion of antagonistic activity was found among the isolates from sponges such as, S. officinalis and D. nigra. Among the bacterial strains, MAPS 15 isolated from S. officinalis exhibited the highest and broadest activity spectrum. The results of agar well diffusion assay with MAPS 15 demonstrate that an extracellular product might be responsible for inhibitory action. The results ensured that sponge-associated microorganisms are highly potential resource of bioactive natural products owing to their competition for nutrition, space and light [15-17]. The results of the invitro assays agree with former studies showing antagonistic activity of bacteria isolated from other sponge species [18]. Scores of studies affirmed that marine bacteria could reduce the growth of pathogenic human and animal bacteria. However, data pertinent to the repression of Vibrios by marine bacteria are limited [8]. Moreover, Vibrios has been associated with food poisoning in human who consumed tainted shell fishes particularly the species such as V. parahaemolyticus and V. vulnificus. The molecular characterization based on partial 16S rRNA sequence revealed that the active isolate MAPS 15 was Streptomyces sp. Species of the genus Streptomyces are well known producers of metabolites with antimicrobial activity [19]. In conclusion, the sponge isolate, Streptomyces sp. exerts significant antagonistic activity against all the tested human and shrimp pathogens. The antagonistic efficacy exhibited by the Streptomyces sp. against pathogens envisaged the future use of sponge-associated bacteria for developing human and veterinary grade antibiotics.
Competing Interests
The authors have no conflicts of interest to disclose.
4749
References
- Manilal A, Sujith S, Selvin J, Shakir C, Gandhimathi R, et al. (2010) Antimicrobial potential ofmarine organisms collected from southwest coast of India against multiresistant human and shrimp pathogens. Sci Mar 74: 287-296.
- Xiong ZQ,Wang JF, Hao YY,Wang Y (2013) Recent Advances in the Discovery and Development of Marine Microbial Natural Products. Mar Drugs 11: 700-717.
- Manilal A, Sabarathnam B, Sujith S,Shakir C,Selvin J(2010) Antagonistic potentials of marine sponge associated fungi Aspergillusclavatus MFD15. Asian JMediSci 2: 195-200
- Holler U, Wright AD, Matthee GF, Konig GM, Draeger S, et al. (2000) Fungi from marine sponges: diversity, biological activity and secondary metabolites. Mycol Res 104: 1354-1365.
- Burja AM, Hill RT (2001) Microbial symbionts of the Australian Great Barrier Reef sponge, Candida spongia flabellate. Hydrobiologia 461: 41-47.
- Osinga R, Armstrong E, Burgess GJ, Hoffmann F, Reitner J, et al. (2001) Sponge- microbe associations and their importance for sponge bioprocess engineering. Hydrobiologia 461: 55-62.
- Sujith S, Manilal A, Selvin J, Idhayadhulla A, Kumar RS (2012) Evaluating the antagonistic potential of seaweed-associated marine bacteria collected from the South Indian littoral. Asian J Anim Vet Adv 7:578-567.
- Gandhimathi R, Arunkumar M, Selvin J, Thangavelu T, Sivaramakrishnan S, et al. (2008) Antimicrobial potential of sponge associated marine actinomycetes. JMed Mycol 18: 16-22.
- Manilal A, Sujith S,Selvin J,Kiran G, Shakir C, et al. (2010) Virulence of vibrios isolated from diseased black tiger shrimp PenaeusmonodonFabricius. J World AquacultSoc 41: 332-343.
- EgorovNS(1965)MicrobeantagonistsandBiologicalAssessmentoftheir Antibiotics Activity, Moscow, VysshayaShkola Publishers (in Russia).
- Lalucat J, Bennasar A, Bosch R, García-Valdés E, Palleroni NJ (2006) Biology of Pseudomonas stutzeri.MicrobiolMolBiol Rev 70: 510-547.
- Lechavalier MP, Lechavalier HA (1970) Chemical composition as a criterion in the classification of aerobic actinomycetes. Int J SystBacteriol 20: 435-443.
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J MolBiol 215: 403-410.
- Ivanova EP, Nicolan DV, Yumoto N, Taguchi T, Okamoto K, et al. (1998) Impact of conditions of cultivation and adsorption on antimicrobial activity of marine bacteria. Mar Biol 130: 545-551.
- Burgess JG, Jordan EM, Bregu M, Mearns-Spragg A, Boyd KG (1999) Microbial antagonism: a neglected avenue of natural products research. J Biotechnol 70: 27-32.
- Armstrong E, Yan L, Boyd KG, Wright PC, Burgess JG (2001) The symbiotic role of marine microbes on living surfaces. Hydrobiologia 461: 37-40.
- Selvakumar D, Arun K, Suguna S, Kumar D, Dhevendaran K (2010) Bioactive potential of Streptomyces against fish and shellfish pathogens. Iran J Microbiol 2:157-64.
- Selvakumar D (2010) Marine Streptomyces as a novel source of bioactive substances.World J MicrobiolBiotechnol 26: 2123-2139.










