Rania AA1*, Nsreen MK1, Rasha HEl2 and Mona MA1
1Department of Clinical and Chemical pathology, Beni-Suef University, Beni Suef, Egypt
2Department of Clinical and Chemical pathology, Cairo University, Egypt
- Corresponding Author:
- Rania A Azouz
Department of Clinical and Chemical pathology
Beni-Suef University
Tel: +20822237741; +201009043903
E-mail: Rania_Azouz@med.bsu.edu.eg
Received Date: November 23, 2017; Accepted Date: November 29, 2017; Published Date: December 31, 2017
Citation: Rania AA, Nsreen MK, Rasha HEl, Mona MA (2017) Evaluation for the Novel mecC Methicillin Resistance among Methicillin Resistant Staphylococcal Isolates in two Egyptian University Hospitals. Arch Clin Microbiol. Vol.9 No.1:71 doi:10.4172/1989-8436.100071
Copyright: © 2017 Rania AA, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Keywords
mecC gene; Methicillin resistance; Staphylococci; PBP2a latex
Introduction
Methicillin resistant S. aureus (MRSA) was first described in 1961 collapsing the treatment options for staphylococcal infections as MRSA exhibits resistance to all B lactam antibiotics except 5th generation cephalosporins. Resistance to antibiotics other than ß lactams is common especially in health care associated MRSA (HA-MRSA). The dissemination of MRSA in the community (community associated MRSA [CA-MRSA]) and in animal adapted lineages of S. aureus (livestock associated MRSA [LA-MRSA]), besides the originally described HA-MRSA represents a public health problem and a burden to infectious disease medicine. The genetic principle for methicillin resistance was identified more than 20 years after the first description of MRSA. It was due to the acquisition of mobile genetic element called staphylococcal cassette chromosome that become integrated into the staphylococcal chromosome carrying the gene encoding for resistance [1]. It was thought that the only gene for methicillin resistance in staphylococci is mecA gene that encodes a transpeptidase PBP2a which has low affinity for ß lactams allowing cell wall synthesis to continue in the presence of the antibiotic. In 2011, whole genome sequencing of S. aureus strain LGA251 and other strains that show phenotypically methicillin resistance despite being mecA negative, revealed a novel mecA homolog that share only 69.7% nucleotide identity with mecA. It is carried on novel SCCmec type XI which has new ccr type 8, divergent mecA regulatory genes (mecI and mecR), and no joining region J3 [2]. The protein product (PBP2aLGA251) of mecC gene has 63% amino acid identity to that of mecA gene and it its affinity towards oxacillin is 4 folds that of PBP2a [3].
Reports from European countries accounted the prevalence of mecC-mediated methicillin resistance to be low 0-2.8% among human MRSA isolates in 13 European countries specially Western and Northern Europe. Despite its recent discovery, mecC gene was found to be widely distributed in animals carried by isolates of S. aureus and other staphylococcal species [4-6].
In this study we tried to evaluate the existence of mecC gene in methicillin resistant staphylococcal isolates obtained from two university hospitals in Egypt.
Materials and Methods
Bacterial isolates
A total of 600 sequential samples of methicillin resistant staphylococcal clinical isolates (520 MRSA and 80 MR-CoNS) were collected from clinical microbiology laboratories of two university hospitals in the duration from March 2014 to May 2016. Methicillin sensitive isolates are excluded. Hospitals sharing in this study are presented in Table 1. Methicillin resistant staphylococcal isolates were identified through routine identification and susceptibility testing. Patients’ data were obtained from microbiology request forms.
| Hospital |
City and Country |
Number of isolates |
Wards |
Duration |
| KasrAlainy hospital |
Cairo, Egypt |
300 MRSA |
ICU units |
March 2014 – September 2014 |
| Beni-Suef university hospital |
Beni-Suefgovernrate, upper Egypt |
220 MRSA and 80 MR-CoNS |
Outpatients and different inpatients’ wards |
April 2015 – May 2016 |
Table 1: University hospitals sharing in this study.
Identification and susceptibility testing by disc diffusion
Colonies from different clinical samples on different agar media were identified by morphology, gram stain and catalase reaction, then culture on mannitol and DNase agar. Tube coagulase test was performed when discrepancy between mannitol and DNase results observed. Routine susceptibility testing by disc diffusion was followed including cefoxitin 30 μg (Oxoid, UK) and oxacillin 1ug (Oxoid, UK) according to CLSI on Muller Hinton agar plates [7,8]. All 600 isolates were screened for the discrepant susceptibility profile oxacillin sensitive/cefoxitin resistant.
PBP2a latex agglutination
Discrepant isolates along with randomly selected isolates to have a total of 50 isolates were tested for PBP2a by latex agglutination (Mast group Ltd, UK) according to instructions by the manifacturer.
Polymerase chain reaction (PCR)
Isolates tested for mecA by conventional PCR included isolates tested by PBP2a latex agglutination and randomly selected isolates From Beni-Suef University Hospital which lies in a semirural area. A total of 150 isolates (110 MRSA + 40 MR-CoNS) were tested for mecA by conventional PCR using forward primer (5'- GTA GAA ATG ACT GAA CGT CCG ATA A-3') and reverse primer (5'-CCA ATT CCA CAT TGT TTC GGT CTA A-3') according to McClure et al. [9]. The PCR reaction contained 12.5 μl mastermix (Emerald Amp GT), 1 μl of each primer, and 6 μl of DNA template to reach a total volume of 25 μl. Thermal cycling was adjusted to 5 minute at 94°C for primary denaturation followed by 35 cycles of 30 seconds denaturation at 94°C, 30 seconds at 50°C for annealing, and 30 seconds at 72°C for extension. Finally extension for 7 minutes at 72°C was followed. Agarose gel (1.5%) electrophoresis and ethidium bromide staining was used to separate and visualize the amplified product which was identified at 310 bp using 100 bp DNA ladder. Isolates negative for mecA were tested for mecC by conventional PCR using forward primer (5'- GCT CCT AAT GCT AAT GCA -3') and reverse primer (5'- TAA GCA ATA ATG ACT ACC-3) according to Cuny et al. [10]. Unlike thermal cycle for mecA, annealing temperature for mecC was adjusted to 50°C for 40 seconds while extension and final extension were adjusted to 72°C for 40 seconds and 10 minutes respectively. The amplified product was identified at 304 bp. ATCC 33591 MRSA was used as a positive control in PCR for mecA and positive control for mecC was obtained from Animal Health Research Institute in Giza, Egypt.
Results and Conclusions
Six hundred human clinical cases were included in this study. The gender, mean age, departments sharing and the initial culture site are presented in Table 2. All 600 isolates were screened for the susceptibility profile oxacillin sensitive/cefoxitin resistant by disc diffusion method as an easy, widely used, and cheap method that may predict the probable existence of mecC gene in isolates that exhibit discrepant susceptibility. Figure 1 shows the percentage of discrepant susceptibility among both MRSA and MR-CoNS isolates.
| |
Cairo University Hospital
[300 MRSA isolates No(%)] |
Beni-Suef University Hospital
[220 MRSA No(%) + 80 MR-CoNS No(%)] |
| Sex |
|
|
Male
Female |
180 (60%)
120 (40%) |
153 (51%)
147 (49%) |
| Age groups |
|
|
Neonates
Pediatrics
Adults |
-
-
300 (100%) mean 50 |
22 (7%)
74 (25%)
204 (68%) mean 47 |
| Departments |
|
MRSA |
MR-CoNS |
ICUs
Surgery ward
Medicine ward
Pediatrics ward
Outpatient clinic |
300 (100%)
-
-
-
- |
92 (42%)
12 (5%)
64 (29%)
26 (12%)
26 (12%) |
28 (35%)
1 (1.25%)
9 (11.25%)
34 (42.5)
8 (10%) |
| Sample types |
|
MRSA |
MR-CoNS |
Wound swabs and pus
Urine
Sputum
Blood
CSF
Others |
128 (43%)
17 (6%)
40 (13%)
115 (38%)
-
- |
29 (13%)
52 (24%)
31 (14%)
101 (46%)
4 (2%)
3 (1%) |
1 (1.25%)
5 (6.25%)
-
72 (90%)
2 (2.5%)
- |
Table 2: Descriptive data of patients and bacterial isolates.
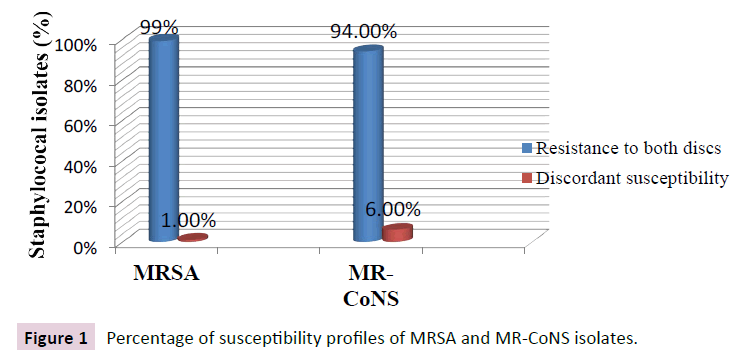
Figure 1: Percentage of susceptibility profiles of MRSA and MR-CoNS isolates.
All discrepant isolates were positive for PBP2a latex agglutination and were also positive for mecA by PCR. Thus discrepant susceptibility by disc diffusion in this study was not due to mecC carriage by any of the isolates. Instead discrepancy was due to the inducible character of oxacillin resistance and heterogeneous expression of methicillin resistance in staphylococci.
Of 150 isolates tested by PCR for mecA gene, 6 isolates tested negative. They were all MRSA isolates (5.5% of MRSA isolates) and were resistant to both cefoxitin and oxacillin by disc diffusion method. Gel electrophoresis for detection of mecA amplicon is shown in Figure 2.
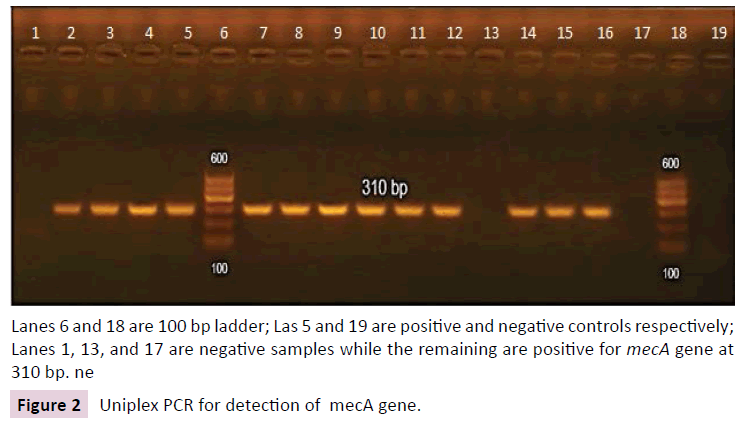
Lanes 6 and 18 are 100 bp ladder; Las 5 and 19 are positive and negative controls respectively; Lanes 1, 13, and 17 are negative samples while the remaining are positive for mecA gene at 310 bp. ne
Figure 2: Uniplex PCR for detection of mecA gene.
All isolates negative for mecA were also negative for mecC by conventional PCR Uniplex PCR for detection of mecC amplicon is shown in Figure 3.

Lane 1: Positive control; Lane 2: 100 bp ladder; Lane 3: Negative control; Lanes 4,5,6,7,8, and 9: Test samples negative for mecC gene
Figure 3: Uniplex PCR for detection of mecC amplicon at 304 bp.
No mecC MRSA isolates were detected in this study. This was in concordance to Ganesan et al. [11] and Basset et al. [12]. Also mecC could not be detected among MR-CoNS isolates in this study. This was the case in Nijjar et al. [13]. The mecA/mecC negative isolates were all positive by cefoxitin screen by Vitek II and all isolates showed oxacillin MIC ≥ 4 μg/ml. This moderately high resistance may exclude Border Line Oxacillin Resistant S. aureus (BORSA) type of resistance to be the cause of methicillin resistance in those isolates. Modified Penicillin binding proteins in S. aureus (MODSA) may be the cause of methicillin resistance in mecA/mecC negative isolates in this study. This needs further investigations as done by Ba et al. who described amino acid substitutions in penicillin binding proteins on whole genome sequencing for MRSA isolates lacking mec genes [14].
In conclusion mecC gene was not detected in any of staphylococcal isolates in this study, though further studies especially in rural areas testing for mec genes in larger number of clinical isolates using multiple primer sets for mecC, may increase the probability of detection of mecC gene or its homologs.
Funding
This work was supported financially by Beni-Suef University.
Ethical Approval
No ethical approval was required for this study as the isolates used in the study were collected as part of routine clinical facility.
Conflict of Interest
The authors state that they have no competing interests.
Informed Consent
Informed consent was not applied, though letters were sent to patients to inform them about the study and who to contact if they did not want to share in the study.
21960
References
- Morand SF, Beaudeau JC (2011) New frontiers of molecular epidemiology of infectious diseases. Springer Science & Business Media 12: 145.
- García-Álvarez L (2011) Meticillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: a descriptive study. The Lancet infectious diseases 11: 595-603.
- Kim C (2012) Properties of a novel PBP2A protein homolog from Staphylococcus aureus strain LGA251 and its contribution to the β-lactam-resistant phenotype. Journal of Biological Chemistry 287: 36854-36863.
- Hiramatu K (2014) Multi-drug-resistant Staphylococcus aureus and future chemotherapy Journal of Infection and Chemotherapy 20: 593-601.
- Becker K (2014) Methicillin resistance in Staphylococcus isolates: the “mec alphabet” with specific consideration of mecC, a mec homolog associated with zoonotic S. aureus lineages. International Journal of Medical Microbiology 304: 794-804.
- Paterson GK, Harrison EM, Holmes MA (2014) The emergence of mecC methicillin-resistant Staphylococcus aureus Trends in Microbiology 22: 42-47.
- CLSI (2013) Performance standards for antimicrobial susceptibility testing.Clinical and Laboratory Standards Institute, Wayne 121.
- McClure JA (2006) Novel multiplex PCR assay for detection of the staphylococcal virulence marker Panton-Valentine leukocidin genes and simultaneous discrimination of methicillin-susceptible from-resistant staphylococci.Journal of clinical microbiology 44: 114.
- Cuny C (2011) Rare occurrence of methicillin-resistant Staphylococcus aureus CC130 with a novel mecA homologue in humans in Germany.PloS one 6: e2436.
- Ganesan A (2013) Evaluation for a novel methicillin resistance (mecC) homologue in methicillin-resistant Staphylococcus aureus isolates obtained from injured military personnel. Journal of clinical microbiology 51: 3073-3075.
- Basset P (2013) Very low prevalence of meticillin-resistant Staphylococcus aureus carrying the mecC gene in western Switzerland. Journal of Hospital Infection 83: 257-259.
- Nijjar CK, Smith MH, Eltringham IJ (2014) Adjunctive mecA PCR for routine detection of methicillin susceptibility in clinical isolates of coagulase-negative staphylococci Journal of clinical microbiology 52: 1678-1681.
- Ba X (2013) Novel mutations in penicillin-binding protein genes in clinical Staphylococcus aureus isolates that are methicillin resistant on susceptibility testing, but lack the mec gene. Journal of Antimicrobial Chemotherapy 69: 594-597.








