Key words
Hepatitis C virus, Laboratory diagnosis, False positivity.
Hepatitis C virus
In the mid-1970s, a new disease entity termed ‘’non-A, non- B’’ (NANB) hepatitis was first described, and in the following years leading to the discovery of the virus, posttransfusion and community-acquired NANB hepatitis increasingly became a potentially serious disease, resulting in cirrhosis and/or hepatocellular carcinoma [1,2]. Hepatitis C virus (HCV) is a singlestranded RNA virus with a genome of about 10.000 nucleotides containing a single large, continuous open reading frame and with an organization most closely resembling the Flaviviridae [3]. Hepatitis C virus infection is a global health problem. The World Health Organization (WHO) estimates that at least 170 million people are infected with HCV worldwide, and most of the patients are concentrated in developing countries [4].
Methods used at laboratory diagnosis of hepatitis C virus infection
Diagnostic procedures of hepatitis C virus infection in laboratories are principally based on the detection of antibodies (IgG) against recombinant HCV polypeptides by two main methods: Enzyme immunoassay (EIA) and Chemiluminescence immunoassay (CIA). Non-structural and recombinant antigens have been used in the production of immunoassay reactives. Serologic and virologic markers of past or present HCV infection include IgG antibodies (anti-HCV). An assay for Ig M anti- HCV is available, but it does not distinguish between acute and chronic HCV infection [5].
Three different generations of HCV kits have been developed to date. The first generation HCV enzyme immunoassay detected only antibodies against non-structural region 4 (NS4) with recombinant antigen c100-3. First-generation anti-HCV reactives had relatively poor specifity and sensitivity. In the development of second-generation tests, additonal antigens from the core region (c22-3), the NS3 region (c33c) and a part of c100-3 from the NS4 region were used. Second-generation reactives show increased sensitivity and specifity. Third-generation EIA includes an additional antigen from the NS5 protein and a reconfiguration of the core and NS3 antigens [6].
Antibodies to HCV are detectable an average of 8 weeks later after an exposure. Antibodies to NS3 and core antigen generally appear a few weeks before the antibodies to NS4 and NS5- antigen. Whereas anti-NS3 is almost always detectable, anticore antibody may initially be absent, especially in the case of self-limiting or asymptomatic infections, and in up to one third of the cases does not appear until the chronic stage. Total serum HCV core antigen, a surrogate marker of HCV replication, can also be detected and quantified and commercial assay is available . HCV core antigen can be detected on average 1 to 2 days after HCV RNA during the pre-seroconversion period. HCV Ag test presented as a ‘’Direct marker for diagnosis of HCV infection’’. Sensitivity of HCV core antigen test is lower than HCV-RNA assay [7, 8].
Diagnostic tests may be divided into three groups:
1- Screening test (anti-HCV tests based on EIA or CIA)
2- Supplemental test (recombinant immunoblot assay-RIBA)
3- Confirmatory test (HCV RNA).
False positivity of anti-HCV tests
Although present tests have better sensitivity and specifity rates than their predecessors, there still exists a high prevelance of false-positive results, especially among low risk group, immunocompromised patients or populations without liver diseases, leading to unnecessary cost-effective health expenditures and confusing diagnostic challenges. The most common problem in the laboratory screening assay of anti-HCV is the false positivity of low titers. Among immunocompetent populations with anti-HCV prevalence less than 10 % (e.g., volunteer blood donors, military personnel, general population, health care workers, or clients attending sexually transmitted diseases clinics ) the proportion of false positive results averages approximately 35 % (range: 15 % to 60 %) [9,10].
Among immunocompromised populations (e.g., hemodialysis patients) average rate of false-positive results approximately: 15 %. False positive anti-HCV results obtained from both CIA and EIA based reactives can be explained with by the fact that no structural antigens and proteins have been derived from HCV up to now. HCV has not to be cultured, natural viral proteins were unlikely obtained. A lot of discussions focused on deciding a reliable, easy-to-use and cost-effective confirmative test method in order to check and confirm the results of anti- HCV reactives routinely used. A confirmatory test is needed to discriminate the false positive results from the accurate ones [11].
Reasons of false positivity
The aminoacid sequence and the purity of the HCV antigen used for assay development are significant factors influencing both the specifity and the sensitivity of anti-HCV immunoassays. Because of the high IgG concentration in human blood (>5 mg/ml)-e.g. in paraproteinemia or auto-antibody production or after Ig G denaturation caused by repeated freezing and thawing or by heat-inactivation of serum samples-there is a strong tendency for some of the IgG molecules to be bound to the micro-well surface by direct adsorbtion or by indirect capture via the surface molecules, and then arouse a signal, giving false-positive results. This problem might be more serious when the samples are from patients with systemic lupus erythematosus (SLE), rheumatoid arthritis (RA), portal cirhosis, and some infectious diseases due to the very complicated, higher concentration of immunoglobulin components in their blood [11].
Supplemental test
RIBA also a kind of serologic assay may be used as a supplemental test. A positive RIBA result interpreted as anti-HCV positive. A confirmed anti-HCV positive result indicates the need for serial determination of HCV-RNA activity. As a result RIBA only may be a confirmative test for the anti-HCV positivity. But it is not enough to confirm current HCV infection. RIBA has several disadvantages, such as the difficulties of performing the test, high percentage of the indetermined results and high cost-effectiveness causing the less use or preventing the common use in many developing countries, in routine diagnostic laboratory procedures [12].
RIBA HCV 3.0 SIA (Strip Immunoblot Assay) contains recombinant antigens and synthetic peptides (Recombinant c33c and NS5 antigens; Synthetic 5-1-1, c100, and c22 peptides). RIBA is positive when patients exhibit HCV RNA positivity. RIBA may be either positive or indetermined or negative when patients exhibit HCV RNA negativity. Mesurements of HCV-RNA should be especially reserved for the patients with indetermined or positive RIBA results. A negative RIBA is indicative of unspecific reactivity in the anti-HCV screening test. A positive RIBA in many cases is not proof of specific reactivity in the screening test [13]. In patients with a high pretest probability of infection, a positive serological screening test should be ‘’confirmed’’ by performing a qualitative test for HCV RNA rather than the supplemental recombinant immunoblot assay. This strategy is cost-effective and more informative than usnig the RIBA to confirm positive antibody screening tests in a diagnostic setting [14].
Confirmation test is needed
Although third-generation HCV reactives are more sensitive and specific than older generation assays, they still have a high percentage of false positive reactions, so that it is mandatory to confirm every reactivity, especially with low titers by anti-HCV CIA or EIA with HCV RNA assay (reactives with a lower limit of detection of 50 IU/mL or less) to avoid false positive results. To minimize the likelihood of false-positive anti-HCV results, CDC has recommended the confirmation of all anti-HCV results by either RIBA or HCV-RNA assay [15,16].
HCV RNA is the earliest marker of infection, and a direct indicator of ongoing viral replication. It appears 1 to 2 weeks after infection before any alterations in liver enzyme levels and appearance of anti-HCV antibodies can be detected. If the nucleic acid testing (NAT) result is positive, active HCV infection is confirmed. If NAT result is negative, the HCV antibody or infection status can not be determined. NAT assays used to detect and quantify HCV RNA: the following target amplification methods used qualitative detection of HCV RNA and have lower limits of detection of 5-50 IU/mL: Reverse Transcriptase-PCR (single/ dual enzyme RT-PCR, nested RT-PCR), TMA (transcription-mediated amplification), NASBA (isothermal RNA amplification). Quantification of HCV-RNA can be determined by comperative or Real-Time PCR assays, signal amplification methods (bDNA) [14,17,18].
In certain situations HCV RNA result can be negative in persons with active HCV infection. As the titer of anti-HCV increases during acute infection, the level of circulating HCV-RNA declines; intermittent HCV RNA positivity has been obsorved among persons with chronic HCV infection. A negative HCV RNA result also can indicate resolved infection. Follow-up HCVRNA testing is indiciated only in persons with serologically confirmed anti-HCV positive results [14,16]. Interpretation of HCV test results presented in Table 1 (2).
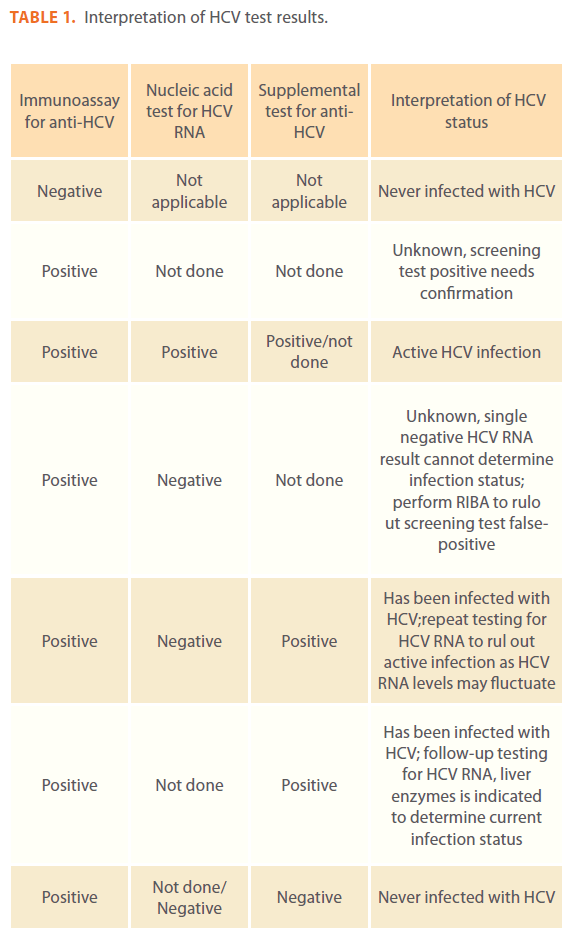
Table 1: Interpretation of HCV test results.
Chronic hepatitis C
Chronic hepatitis C is certain when both anti-HCV antibodies and HCV-RNA are present. Detectable HCV replication in the absence of anti-HCV antibodies is exceptional with the current enzyme immunoassays, almost exclusively observed in: profoundly immunodepressed patients, hemodialysis patients, agammaglobulinemic subjects [19,20].
Conclusion
The diagnosis of HCV should be as reliable as possible, because a positive result has a deep impact on the life of the affected person. Commonly used anti-HCV CIAs or EIAs produce a high percentage of false positive results, proving the neccesity of the use of an easy-to-use, reliable and cost-effective confirmation test, HCV-RNA, in routine diagnostic laboratory procedures.
193
References
- Feinstone SM, Kapikian AZ, Purcell RH, Alter HJ, Holland PV (1975) Transfusion-associated hepatitis not due to viral hepatitis type A or B. N Eng J Med 292:776-770.
- Lemon SM, Walker C, Alter MJ, Yi MK. (2007) Hepatitis C virus In: Knipe DM, Howley PM editors-in chief. Fields Virology.5th ed. Wolters Kluwer Lippincott Williams & Wilkins, Philadelphia, USA. pp.1253-1304.
- Choo Q-L, Richman KH, Han JH, Berger K, Lee C, et al. (1991) Genetic organization and diversity of the hepatitis C virus. ProcNatlAcadSci USA 88:2451-2455.
- Global surveillence and control of hepatitis C. (1999) Report of a WHO Consultation organized in collaboration with the Viral Hepatitis Prevention Board, Antwerp, Belgium. J Viral Hepat 6:35-47.
- Kuo G, Choo QL, Alter HJ, Gitnick GL, Redeker AG, et al. (1989) An assay for circulating antibodies to a major etiologic virus of human non-A, non-B hepatitis. Science 244:362-364.
- Feucht HH, Zöllner B, Polywka S, Laufs R. (1995) Study on reliability of commercially avilaible hepatitis C virus antibody tests. J ClinMicrobiol 33: 620-624.
- Frösner G. Viral hepatitis. (1998): In Thomas L. editor. CLinical Laboratory Diagnostics.TH-Books, Frankfurt, Germany. pp.1260- 1287.
- Colin C, Lanoir D, Touzet S, Meyaud-Kraemer L, Bailly F, et al. (2001) Sensitivity and specifity of third-generation hepatitis C virus antibody detection assay: an analysis of the literature. J Viral Hepat 8:87-95.
- Ansari MHK, Omrani MD. (2006) Evalutaions of diagnostic value of ELISA method (EIA) & PCR in diagnosis of hepatitis C virus in hemodialysis patients. Hepatitis Monthly 6:19-23.
- Centers for Disease Control and Prevention. (2003) Guidelines for laboratory testing and result reporting of antibody to hapatitis C virus. MMWR Morbid Mortal Wkly Rep 52:1-13.
- Wu FB, Ouyan HQ, Tang XY, Zhou ZX (2008) Double-antigen sandwich time-resolved immunoflourometric assay for the detection of anti-hepatitis C virus total antibodies with improved specifity and sensitivity. J Medical Microbiol 57:1-7.
- Kesli R, Ozdemir M, Kurtoglu MG, Baykan M, Baysal B (2009) Evaluation and comparison of three different anti-HCV reactives based on chemiluminescence and enzyme immunoassay method used in the diagnosis of Hepatitis C ?nfections in Turkey. J Int Med Res 37: 1420-1429.
- Dufor DR, Talastas M, Fernandez MDA et al. (2003) An important predictor factor of low likelihood of hepatitis C infection. Clinical Chemistry 49:479-486
- Fiebelkorn KR, Nolte SH. (2004) RNA virus detection. In Persing DH editor in chief. Molecular Microbiology Diagnostic Principles and Practise. ASM Press, Washington DC, USA. pp.441-474.
- Schröter M, Feucht HH, Schafer P, Zöllner B, Polywka S, et al. (1999). Definition of false-positive reactions in screening for hepatitis C virus antibodies. J ClinMicrobiol 37:233-234.
- Chevaliez S, Pawlotsky JM (2006). Hepatitis C virus serologic and virologic tests and clinical diagnosis of HCV-related liver disease. Int J Med Sci 3:35-40.
- Chevaliez S, Pawlotsky JM (2005) Use of virologic assays in the diagnosis and management of hepatitis C virus infection. Clin Liver Dis 9:371-382.
- Saldanha J, Lelie N, Heath A, and WHO Collaborative Study Grouop. (1999) Estabilishment of the first international standart for nucleic acid amplification technology (NAT) assays for HCV-RNA. Vox Sang 76:149-158.
- Strader DB, Wright T, Thomas DL, Seeff LB, (2004) Diagnosis Management, and treatment of hepatitis C. Hepatology 39:1147- 1171.
- Chevaliez S, Pawlotsky JM. (2007) Hepatitis C virus: Virology, diagnosis and management of antiviral therapy. World Gastroenterol 13:2461-2466.






