Introduction
Resistant bacteria are emerging worldwide as a threat to the favourable treatment outcome of common infections in community and hospital settings [1]. Among antibiotics ,β- lactams are the most widely used agents accounting for over 50 % of all systemic antibiotics in use [2]. Mechanisms by which clinical isolate of Gram negative bacteria resist β-Lactam antibiotics are through production of β–Lactamases, modification of cell wall and modification of target sites with reduced affinity for β-Lactam antibiotics [3-4]. Among these, the production of β-lactamase appears to be of primary concern and one of the most rapidly developing and clinically significant antimicrobial resistance mechanism [3-4].
The first plasmid mediated β-lactamase in Gram negatives was reported in 1965 from an E. coli isolate from a patient in Athens, Greece, named Temnoniera (hence the designation TEM.). Another common plasmid mediated β-lactamase found in Klebsiella pneumonia (K.pneumoniae.) and E. coli is SHV-1 (named after the sulfhydryl “variable”active site) [5]. Newer β-lactam antibiotics (extended spectrum βeta-lactam) that would not be susceptible to these enzymes became widely used in the 1980’s [3]. The first report of plasmid-encoded β-lactamase capable of hydrolysing the extended spectrum cephalosporins was published in 1983[5] which was first isolated in Germany. ESBLs spread rapidly to Europe, US and Asia and are now found all over the world [2]. Being plasmid mediated, they are easily transmitted among members of enterobacteriaceae thus facilitating the dissemination of resistance not only to β-lactams but to other commonly used antibiotics such as quinolones and aminoglycosides [2].
E. coli is one of the most common isolate in our hospital settings and as β-lactam antibiotics are mainstay of treatment, the increasing number of E .coli isolates exhibit ESBLs; and as such β-lactam group of antibiotics will be almost ineffective in few years to come. E. coli is most common organism after Klebsiella to exhibit ESBL’s. There is paucity of information about antimicrobial resistance especially on ESBL’s from Kashmir. The present study was undertaken, keeping in view of the above facts, to know the prevalence and resistance patterns of clinical isolates of ESBL producing E. coli by employing DDST and PCDDT. In addition, E-test method of detection was performed on selected ESBL positive strain.
Methods
Clinical isolates
A total of 221 E. coli isolates from different clinical specimens during the period of August 2005 to July 2007, were screened for potential ESBL activity. These strains were isolated from different clinical specimens like urine, blood, sputum, pus and other body fluids which were received in the bacteriological division of microbiology. All the samples were processed and identified as per the standard bacteriological division of microbiological protocols and procedures [6]. E .coli ATCC 25922 was used as a negative ESBL control. Based on routine antibiotic disc sensitivity tests, isolates that exhibited resistance to any one of the third generation cephalosporins: ceftazidime, cefotaxime, ceftriaxone were shortlisted to test for the production of ESBL.
Antibiotics
The following antibiotic sensitivity discs were used for primary screening: ceftazidime 30 μg, cefotaxime 30 μg and ceftriaxone 30 μg. In addition, Augmentin disk containing 20 μg of amoxicillin plus 10 μg of clavulanic acid and ceftazidime (30 μg) + cavulanic acid (10 μg) were used for confirmatory tests. E-test strips of ceftazidime and ceftazidime + clavulanic acid were used for selected ESBL isolates.
Screening for ESBLs by DDST: [7,9-14]
E. coli isolates that exhibited resistance to third generation cephalosporins were screened to detect ESBL producers. Cefotaxime 30 μg disc was placed at a distance of 15 mm edge to edge from a centrally placed disc containing 20 μg of amoxicillin + 10 μg of clavulanic acid. Plates were incubated at 35°C for 18-20 hours and the pattern of zones of inhibition was noted. Isolates that exhibited a distinct shape/size with potentiating towards amoxicillin + clavulanic disc were considered potential ESBL producers.
PCDDT: [7, 9-11]
ESBL production was confirmed among potential ESBL producing isolates by phenotypic tests. Third generation cephalosporins with and without clavulanic acid were used as follows;- ceftazidime (30 μg) and ceftazidime (30 μg) +clavulanic acid (10 μg). Disk diffusion assay was carried out as per guidelines of Clinical Laboratory Standards Institute (CLSI), 2005 and difference in zone diameters between disk with and without clavulanic acid were recorded [29]. A difference of >5 mm between the zone diameters of ceftazidime and ceftazidime + clavulanic acid disk is taken to be phenotypic confirmation of ESBL production.
E-test of selected ESBL isolates. [7-8]
E-test of selected ESBL isolates was carried out. Ratio of ceftazidime MIC and ceftazidime + clavulanic acid MIC equal to or greater than 8 indicated the presence of ESBL.
Results
Two hundred and eleven of 221 isolate (95.5 %) were positive for potential ESBL producers and these isolates were further subjected to confirmatory tests. One hundred and eighteen E. coli isolates tested positive for ESBL production by three cofirmatory tests ; DDST, PCDDT and E-Test. Double disk potentiation test showed eleven E.coli isolates positive for ESBL production. i.e.; 9.3% (11/118) as indicated in figure 1. PCDDT detected 99.2% (117/118) E. coli positive for ESBL production (figure 1). Ten isolates among 117 potential ESBL producers were also confirmed by DDST. Twenty selected isolates were confirmed by E-test method. All of them tested positive for ESBL production with MIC’s ranging from 42.66 μg/ml to 320 μg/ml (Figure 1).
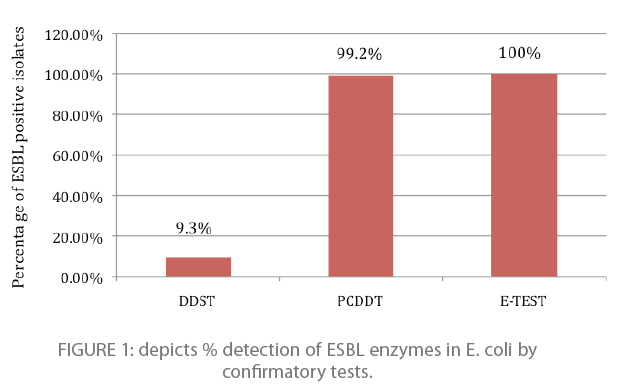
Figure 1: depicts % detection of ESBL enzymes in E. coli by confirmatory tests.
The highest number of ESBL positive isolates were from in patients (71.2 %) followed by (28.8 %) from outpatient (Table 1). The distribution of ESBL positive isolates among wards was as follows; 11.8 % from nephrology, 8.4 % from gastroenterology and general medicine each while the least number was isolated from haematology 1.7 % (Table 1).
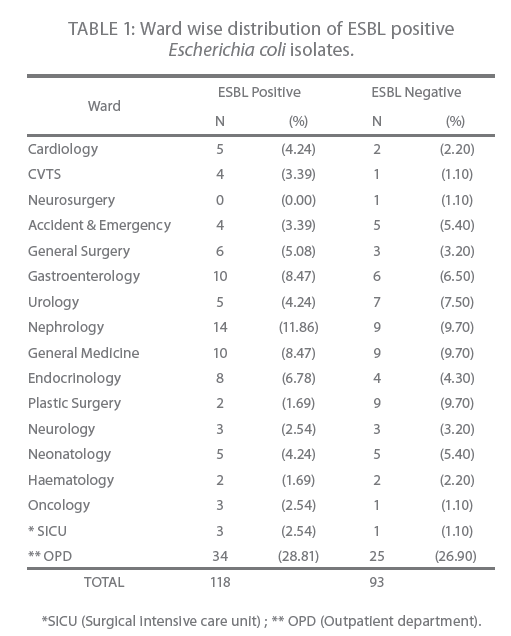
Table 1: Ward wise distribution of ESBL positive Escherichia coli isolates.
Urine (72.9 %) was the main source of ESBL producing isolates followed by pus (9.3 %) and then blood (7.6 %) (Table 2).
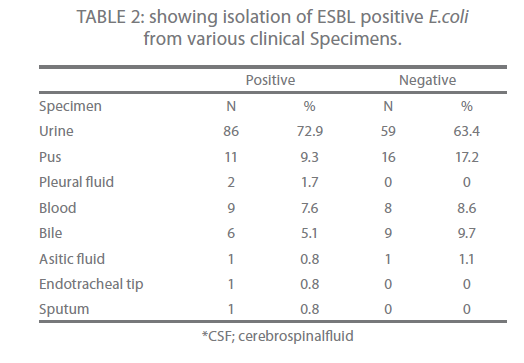
Table 2: showing isolation of ESBL positive E.coli from various clinical Specimens.
The prevalence of ESBL producing E.coli among potential producers of ESBL was 55.9 % (118/211) which was confirmed by DDST, PCDDT and E-test.
Third generation cephalosporins showed 97.5 % to 99.2 % resistance while cephalosporin + clavulanic acid combination reported only 20 % resistance in ESBL producers (Table 3). Fourth generation cephalosporin, cefpime showed 100% resistance. In quinolones; levofloxacin and moxifloxacin reported 100 % resistance followed by ofloxacin and ciprofloxacin 96.2 % and 93.1 % respectively. In aminoglycosides, gentamicin showed 65.2 % resistance. Cotrimaxozole resistance was seen in 69.1 % isolates. Imipenem, nitrofurantoin, meropenem, gatifloxacin and amikacin showed 98.3 %, 91.5 %, 88.9 %, 64.1 % and 78.2 % sensitivity respectively (Table 3).
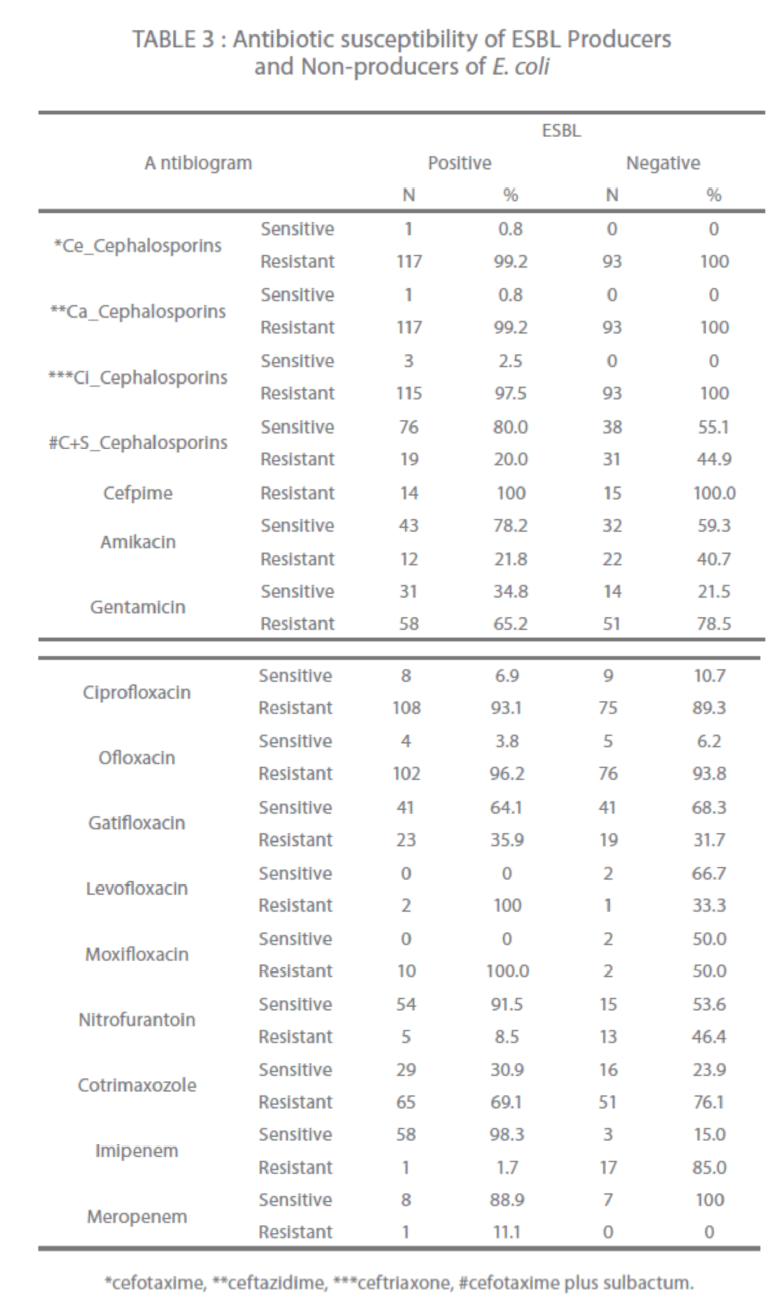
Table 3: Antibiotic susceptibility of ESBL Producers and Non-producers of E. coli
In Non-ESBL producers 100 % resistance seen among third and fourth generation cephalosporins while in quinolones; ciprofloxacin had 89.3 % and ofloxacin 93.8 % resistance. In aminoglycosides, gentamicin had 78.5 % resistance. Cotrimaxozole had 76.1 % resistance (Table 3).
Discussion
ESBLs are clinically important because they destroy cephalosporins, given as first line agents to many severely ill patients. Delayed recognition and inappropriate treatment of severe infections caused by ESBL producers with cephalosporins has been associated with increased mortality [8,30].
In this study, 95.5 % of the E. coli isolates were positive using cefotaxime, ceftazidime and ceftriaxone 30 μg each discs as initial screening agent which were further subjected to confirmatory tests .The high percentage of resistance was seen to 3rd and 4th generation cephalosporins; ceftazidime (99.2 %), cefotaxime (99.2 %), ceftriaxone (97.5 %) and cefpime (100 %) and these findings matched with various studies conducted in India and elsewhere [3,18-19].
The 9.3 % ESBL positive strains confirmed by DDST is very much low when compared to other study [3]. The reasons for discordance are various factors like precise placement of discs, correct storage of the clavulanic discs and performance of appropriate control tests are critical to the sensitivity of DDST. Double disk test can lack sensitivity because of the problems of optimal discs spacing [3,9,11] but our study result was comparable with a study conducted in a teritiary care at Medical College of Virginia [14]. PCDDT result obtained is similar to what has been found in other studies [11,20]. E-test confirmation test method detected ESBL production in twenty selected ESBL positive isolates confirmed by other confirmatory tests. Their ESBL status and MIC was determined and comparable with other studies done [21-22].
The highest number of ESBL positive isolates were from inpatients followed by outpatients which matched with a study reporting ESBL producing bacteria 87 % from inpatients and 12.7 % from outpatients [23].This high percentage of ESBLs from outpatient should alert the physician. Urine was the main source of ESBL’s production in all the specimen followed by pus and blood which was almost similar to study by Bithikia et al [3]and Rafay et al [18]. Much higher (58 %) prevalence of ESBL producers in urinary isolates of gram negative bacilli was observed in India by Mathur et al [24].The out-patient presence of ESBL is of main concern as it is now come to the alert of the physician that ESBL is spreading fast in the community and responsible for communityacquired ESBLs and the highest number being from urine specimens [1,10-11].
The prevalence of ESBL positive E. coli of 55.9 %(118/211) was confirmed by DDST, PCDDT and E-test .The above prevalence of ESBL positive isolates is in concordance with various studies and vary among different geographical areas, countries and institutions [3,19,24,15].
Amit-Jain et al [19] detected marked geographic variation. Incidence in India was 47.5 % and other countries 36 % to 55 %. A study from Northern India [19] in 2000 by Amit Jain reported an incidence of 58.06 % for ESBL producing E. coli which is almost equal to our current study conducted.
The frequency of ESBL-producing organisms differ significantly in accordance with geographic location. Although frequency of ESBL producing E.coli in Europe, North Latin America and Western Pacific is reported at 1-8 %, its prevalence in the Asia Pacific region and South Africa is reported at more than 20 % [25]. The lower prevalence of ESBL in western countries compared to others can be explained by implementation of infection control measures, restricted and judicious use of oxyimino-cephalosporins, implementation of appropriate ESBL detection methods recommended by CLSI, importance of hand hygiene reinforcement, recommended isolation precautions for patients colonized or infected with ESBL producers, continuous education programs and research studies which all are known to contribute to decreasing the spread of ESBL producing organisms.
The antibiogram of both β-lactam and non-β-lactam group of antibiotics showed multidrug resistance pattern (Table 3). A study by Tankhiwale [26] reported 82 % resistance to cotrimaxozole. Nitrofurantoin constituted a reasonable option for treatment (62.5 % sensitivity). Amikacin was drug of choice as 85.7 %isolates were sensitive to this drug. Both these drugs showed similar results in our study.
Cephalosporin plus sulbactam showed 20 % resistance which was higher than reported in a study [16].A study by Villanvena [9] reported multidrug resistance pattern similar to our study but reported 100 % sensitivity to imipenem and meropenem in contrast to our study showing 11.1 % resistance in meropenem and 1.7 % resistance to imipenem as such these two drugs can no longer be considered a drug of choice for ESBL producing E.coli and further these finding corroborated with other studies [16,18,23-24,26].
ESBL producing organisms hydrolyse β-lactam antibiotics, so antibiotic choice for infections with such organism is seriously reduced. Further, the plasmids bearing the genes encoding ESBLs frequently also carry genes encoding resistance to aminoglycosi- des and trimethoprim/ sulfamethoxazole [27].
In the current study multidrug resistance is observed to 3rd and 4th generation cephalosporins, quinolones and aminoglycosides. ESBLmediated resistance is not always obvious in vitro to all cephalosporins. Many ESBL producers are multiresistant to non-β-lactam antibiotics such as quinolones and aminoglycosides, thereby narrowing treatment options. Some producers achieve outbreak status spreading among patients and locals, perhaps owing to particular pathogenicity traits [30]. Once an ESBLproducing strain is detected, the laboratory should report it as “resistant” to all pencillins, cephalosporins and aztreonam, even if they test as susceptible. Other anti-microbial agents can be reported as they are tested [28].
The high prevalence of ESBL in E.coli exists in our institute can be traced to the indiscriminate use of 3rd and 4th generation cephalosporins. The presence of ESBL in outpatient is of main concern as it can be responsible for spreading ESBL from hospital to general population. Therfore, judicious use of 3rd and 4th generation cephalosporins will be effective means of controlling and decreasing the spread of ESBL.
270
References
- Chaudhary U, Aggarwal R (2004) Extended spectrum β-lactamases (ESBL)-an emerging threat to clinical therapeutics. Indian J Med Microbiol 22(2):75-80.
- Kumar MS, Lakshmi V, Rajagopalan R (2006) Occurrence of extended spectrum β-Lactamases among enterobactereaceae spp. isolated at a tertiary care institute. Indian J Med Microbiol 24(3):208-11.
- Duttaroy B, Mehta S (2005) Extended spectrum β-lactamases (ESBL) in clinical isolates of Klebsiellapneumoniaeand Escherichia coli. Indian J PatholMicrobiol 48(1):45-48.
- Ghatole M, Manthalkar P, Kandle S, Yemul V, Jahagirdar V (2004) Correlation of extended spectrum β-lactamases production with Cephalosporin resistance in gram negative bacilli. Indian J PatholMicrobiol 47(1):82-84.
- Al-Jasser A M (2006) Extended-spectrum β-lactamases (ESBL’s):Globalproblem.K M J 38(3):171-85.
- Collee J G, Miles R S,Walt B (1999) Tests for the identification of bacteria in ‘Mackie and McCartney as editors. Practical Medical Microbiology, Ed. fourteenth, Elsiver. Pub Churchill Hill Livingstone, pp. 131-50
- D’Azevedo P A,Goncalves A L S,Musskopf M I,Ramos C G,Dias C A.G. (2004) Laboratory tests in the detection of extended spectrum β-Lactamase production: National Committee for Clinical Laboratory Standards (NCCLS) screening test, the E-test, the double disk confirmatory list, and cefoxitin susceptibility testing. Braz J Infect Dis. 8(5):372-77.
- Villanueva F D,Tupasi T E,Abiad H G, Baella B Q Candano R C (2003) Extended-spectrum β-lactamase production among Escherichia Coli and Klebsiella spp. at the Makat Medical Centre: Tentative Solutions. Phil J Microbiol Infect Dis 32(3):103-08.
- Jenny Andrews. Clinical scientist.Detection of extended-spectrum β-Lactamase (ESBLs) in E.coliand Klebsiellaspecies.BSAC, British society for antimicrobial chemotherapy. https://www.bsac.org.uk/_db/_documents/Ecoliklebsiella.pdf. Accessed 21 Ju1y 2010.
- Luzzano F, Mezzatesta M, Li C M, Perilli M, Stefani S, Amicosante G, Rossolini G M, et al. (2006) Trends in production of extended-spectrum β-Lactamases among enterobacteria of medical interest: Report of the Second Italian Nationwide Survey. J ClinMicrobiol 44(5):1659-64.
- Andrea J.Linscott and William J.Brown (2005) Evaluation of Four commercially available extended-spectrum β-Lactamase phenotypic confirmatory tests. J ClinMicrobiol 43(3):1081-85.
- Vercauteren E, Descheemaeker P, IevenM,.Sanders C C, Goossens H (1997) Comparison of screening methods for detection of extended-spectrum β-Lactamases and their prevalence among blood isolates of Escherichia coli and Klebsiellaspp. In a Balgian Teaching Hospital. J ClinMicrobiol 35(9): 2191-97.
- Shan Muganathan C, Anantha Krishnan A, Jaya Keerthi SR, Kanungo R, Kumar A, Bhattacharya S, et al. (2004) Learning from an outbreak: ESBL- The essential points. Indian J Med Microbiol 22(4):255-57.
- Emery C L,Weymouth L A (1997) Detection and clinical significance of extended spectrum β-Lactamases in a tertiary care medical centre. J ClinMicrobiol 35(8):2061-67.
- Ali A.M, Rafi S, QureshiA.H (2004) Frequency of extended spectrum β-Lactamase producing gram negative bacilli among clinical isolates at clinical laboraties of Army Medical College, Rawalpindi. J Ayub Med CollAbottabad.16(1)135-7.
- Yunsong Y, Weilin Z, Yagang C, Xiang Y, YilinM.A (2002) Epidemiological and antibiotic resistant study on extended-spectrum βeta-lactamase producing Escherichia coli and Klebsiellapneumoniaein Zhejiang Province. Chin Med J 115(10):1479-82
- Ahmed I,Salam A (2002) Extended spectrum βeta-lactamases and bacterial resistance. Pak J Med Sci 18(2):151-55
- Rafay A M, Al-Muharrmi Z, Toki R (2007) Prevalence of extended spectrum β-Lactamases producing isolates over a 1 year period at a University Hospital in Oman. Saudi Med J 28(1):22-27.
- Jain A, Roy I, Gupta M K, Kumar M, Agarwal S K (2003) Prevalence of Extended Spectrum β-Lactamases producing gram negative bacterial septicaemic neonates in a tertiary care hospital. J Med Microbiol 52:421-425.
- M’ZaliFH, Chanawong A, Kerr K G, Birkenhead D, Hawkey P M. (2000) Detection of extended-spectrum β-Lactamases in members of the family Enterobacteriaceae: Comparison of the MAST DD test, the double disc and the E-test ESBL. J AntimicrobChemother, 45:881-85.
- Rodrigues C, Joshi P,.JainS.H, Alphonse M, Krishnan R.R,.Mehta A, (2004) Detection of β-Lactamases in Nosocomial gram negative clinical isolates. Indian J Med Microbiol 22(4):247-250.
- Tenover FC, Mohammed MJ, Gorton T S, Dembek Z F (1999) Detection and Reporting of organisms producing extended-spectrum β-Lactamases: Survey of laboratories in Connecticut. J ClinMicrobiol37:(12): 4065-70
- Kader A A., Angamuthu K (2005) Extended-spectrum β-Lactamases in urinary isolates of Escherichia coli, Klebsiellapneumoniaeand other gram-negative bacteria in a hospital in Eastern Province, Saudi Arabia. Saudi Med J 26(6):956-59.
- Padmini S B, Appalaraju B (2004) Extended Spectrum β-Lactamases in urinary isolates of E.coliand Klebsiella pneumonia prevalence and subsceptibility pattern in a tertiary care hospital. Indian J Med Microbiol 22(3):172-74.
- Jabeen K, Zafar A, Hasan R (2005) Frequency and sensitivity pattern of extended spectrum β-Lactamase producing isolates in a tertiary care hospital laboratory of Pakistan. J Pak Med Assoc. 55(10):436-39.
- Tankhiwala S S,Jalgaonkar S V, Ahamad S , Hassani U (2004) Evaluation of extended spectrum β-Lactamase in urinary isolates. Indian J Med Res 120:553-56
- Paterson D L,Bonomo R A (2005) Extended spectrum β-Lactamases: A clinical update. ClinMicrobiolRev.18(4):657-86.
- Samaha-Kfoury J N, Araj G F (2003) Recent developments in β-Lactamases and extended spectrum β-Lactamases. B M J 327(7425):1209-13.
- Clinical and Laboratory Standards Institute (2005) Performance Standards for antimicrobial susceptibility testing: 15th informational supplement. M 100-S15.Clinical and Laboratory Standards Institute,Wayne.Pa.
- Livermore D M, Ford N W (2005) Laboratory detection and reporting of bacteria with extended spectrum b-Lactamases. Issue No. 2, Issue date 13.06.05. Issued by Standard Unit,Evaluation and Standards Laboratory, Page 1 of 15, Reference No.QS0P5112.www. evaluations-standards.org.uk.Accessed 21 July 2010.









