Nyasha Chin’ombe1*, Ellen Munemo2, Marcelyn Magwenzi1, Boniface Muzividzi2, Pasipanodya Nziramasanga1
Department of Medical Microbiology,College of Health Sciences, University of Zimbabwe, Avondale, Harare, Zimbabwe
National Microbiology Reference Laboratory, Harare, Zimbabwe
- *Corresponding Author:
- Nyasha Chin’ombei, PhD
Department of Medical Microbiology, College of Health Sciences
University of Zimbabwe, Avondale, Harare, Zimbabwe
Tel: +2634307757
E-mail: nyasha.chinombe@gmail.com
Received date: October 30, 2015, Accepted date: Novemebr 16, 2015, Published date: Novemebr 26, 2015
Mycobacterium elephantis is a Nontuberculous Mycobacterium (NTM) and was originally discovered in an elephant that had died of a respiratory disease. This Mycobacterium species was later found in human clinical specimens in a number of countries. In Zimbabwe, M. elephantis was not previously reported. This report is the first one. Using 16S ribosequencing, we discovered one case of M. elephantis in cow dung and two cases in human sputum samples collected in Zimbabwe. In all three cases, the 16S ribosomal RNA gene sequences were 100% identical to the M. elephantis sequences found in the Genbank. The sources of human infection by M. elephantis in Zimbabwe were not clear. Further studies are therefore necessary to ascertain the prevalence and clinical relevance of M. elephantis in humans in Zimbabwe.
Keywords
Mycobacterium elephantis, cases, ribosequencing, Zimbabwe.
Introduction
Mycobacterium elephantis is a NTM that was originally isolated from an elephant which had died of chronic respiratory infection in Sri Lanka in 2000 [1]. Since then, several reports of M. elephantis infection in humans have been published. M. elephantis was isolated in humans in Asia, Belgium, Italy, Canada and Turkey [2-6]. In Africa, M. elephantis in humans was reported recently in Zambia [7]. In most reported cases, the infected humans were sick and had respiratory disease symptoms. Recently, a draft genome for M. elephantis was successfully sequenced [8]. In Zimbabwe, no previous studies have been reported on the identification of M. elephantis in animals or humans. The objective of this study was to report the isolation of M. elephantis from cow dung and human sputum samples in Zimbabwe.
Methods
In 2014, we began isolating Mycobacteria from cow dung samples collected throughout Zimbabwe [9]. During the same time, there was also a national Tuberculosis (TB) survey in Zimbabwe in which 961 nontuberculous Mycobacterium isolates were made. DNA mycobacterial isolates was isolated as previously described [9]. The 5'-end hypervariable region of the 16S ribosomal RNA gene of Mycobacterium was amplified using polymerase chain reaction from the extracted DNA as previously described [9]. The Mycobacterium-specific forward primer, 5′-CCT GCA CTT CGG GAT AAG CCT G -3′, and reverse primer, 5′-CAA CGC GAC AAA CCA CCT ACG A -5′ were used in the polymerase chain reaction. The following cycling program was used for amplification: initial denaturation of 5 minutes at 95°C followed by 35 amplification cycles of 30 seconds at 95°C, 30 seconds at 55°C, and 45 seconds at 72°C, and ending with a final extension step of 7 minutes at 72°C. The amplicons were analyzed by 2% agarose gel electrophoresis. DNA Sequencing of 26 amplicons from cow dung isolates and 81 amplicons from human sputum isolates was performed at Inqaba Biotech (South Africa) using standard protocols. Sequence data were analyzed using Geneious Basic program (Biomatters, USA) and Basic Local Alignment Search Tool (BLAST) programs from the internet. Mycobacterium species were identified using these software programs by comparison of the alignment with known sequences in the Genbanks.
Results
From cow dung samples, 26 isolates of Mycobacteria were identified. Out of these, 1 sample was identified as Mycobacterium elephantis. From the 81 Mycobacterium isolates from human sputum, 2 were identified as Mycobacterium elephantis. The ribosomal DNA sequence (Figure 1) of the three isolates was 100% identical. The sequence was also 100% identical to 6 M. elephantis sequences from Genbanks (Accession numbers FJ497247, AF385898, GQ924944, GU142921, NR_025296, AJ536100) (Figure 2). Sequence with Accession number FJ497247 is from the original M. elephantis isolate from the infected Sri Lanka elephant. AF385898 isolate was from human sputum sample in Canada, GQ924944 isolate was from human sample in Greece and GU142921 from the first human isolate in Asia. AJ536100 was isolated from human sample in Italy and the sequence differed by others by only one nucleotide base. This sequence was identical with two sequences of M. pulveris, a vey close relative M. elephantis (Figure 2).

Figure 1: Nucleotide sequence of 16S rRNA gene of 3 Mycobacterium elephantis isolates discovered in Zimbabwe. Only the 5′-end of the 16S rRNA gene that is hypervarible across Mycobacterium species was amplified, sequenced and analyzed.
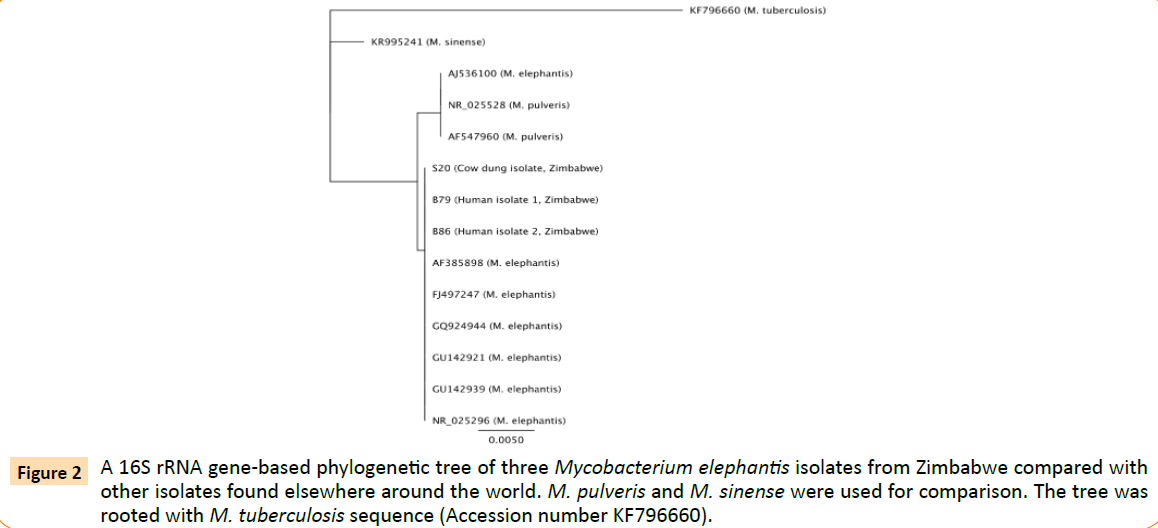
Figure 2: A 16S rRNA gene-based phylogenetic tree of three Mycobacterium elephantis isolates from Zimbabwe compared with other isolates found elsewhere around the world. M. pulveris and M. sinense were used for comparison. The tree was rooted with M. tuberculosis sequence (Accession number KF796660).
Discussion
Molecular identification of microorganisms is now becoming an important component in the diagnosis and treatment of infectious diseases such as tuberculosis and non-tuberculosis. Amplification by polymerase chain reaction and sequencing of 16S ribosomal RNA gene is now being used in the rapid identification of bacterial microorganisms [10]. In this study, we employed 16S ribosequencing in identifying Mycobacterium species in cattle and humans. From the study, three cases of M. elephantis were discovered. This is the first report on the isolation of M. elephantis species in cow dung and humans in Zimbabwe. NTM such as M. elephantis have clinical importance to low-resource countries such as Zimbabwe. These bacteria which are found in diverse environmental sources such as natural and municipal waters, soil, birds and animals can potentially cause opportunistic infections in humans especially those with underlying medical conditions such as HIV/AIDS [11,6]. The source of human infection by M. elephantis in Zimbabwe is still not clear. If the pathogen is present in Zimbabwean elephants, which are many, it can easily move to humans through zoonotic transmission. There is an overlap of elephant, cattle and human ecology in Zimbabwe. Cattle can get infected from elephants and they can potentially transmit the pathogen to humans. In Zimbabwe, humans also live in close proximity with animals and they can easily get infected. The consumption of animal products such as milk and meat can be another potential source of infection. Therefore further studies are necessary to establish the true prevalence and clinical relevance of M. elephantis in Zimbabwe.
Conflicts of interest
The authors declare that there is no conflict of interests.
Acknowledgements
The authors acknowledge funding from the University of Zimbabwe Research Board and the National Microbiology Reference Laboratory of Zimbabwe.
7615
References
- Shojaei H, Magee JG, Freeman R, Yates M, Horadagoda NU, et al. (2000) Mycobacterium elephantis sp. nov., a rapidly growing nonchromogenic mycobacterium isolated from an elephant. International Journal of Systematic and Evolutionary Microbiology 50: 1817-1820
- Heidarieh P, Shojaei H, HashemiA,Feizabadi MM, Daei-Naser A, et al. (2011) First report of isolation of Mycobacterium elephantis from bronchial lavage of a patient in Asia. Journal of the Royal Society of Medicine Short Reports2: 26
- Tortoli E, Rindi L, Bartoloni A, Garzelli C, Mantella A, et al. (2003) Mycobacterium elephantis: not an exceptional finding in clinical specimens. European Journal of Clinical Microbiology and Infectious Diseases 22: 427-430
- Potters D, Seghers M, Muyldermans G, PierardD, Naessens A, et al. (2003) Recovery of Mycobacterium elephantis from sputum of a patient in Belgium. Journal of Clinical Microbiology 41: 1344
- Gunaydin M, Yanik K, Eroglu C, Sanic A, Ceyhan I, et al. (2013) Distribution of nontuberculous Mycobacteria strains. Annals of Clinical Microbiology and Antimicrobials 12:33
- Turenne C, Chedore P,Wolfe J, Jamieson F, May K, et al.(2002) Phenotypic and Molecular Characterization of Clinical Isolates of Mycobacterium elephantis from Human Specimens. Journal of Clinical Microbiology 40: 1230-1236
- Buijtels PC, van der Sande MA, Parkinson S, Verbrugh HA, Petit PL, et al. (2010) Isolation of non-tuberculous mycobacteria at three rural settings in Zambia; a pilot study. Clinical Microbiology and Infection 16: 1142-1148
- Greninger AL, Cunningham G, Yu JM, Hsu ED, Chiu CY et al. (2015) Draft Genome Sequence of Mycobacterium elephantis Strain Lipa.Genome Announcements 3: e00691-e00615
- Padya L,Chin'ombe N, Magwenzi M, Mbanga J, RuhanyaV et al.(2015) Molecular Identification of Mycobacterium Species of Public Health Importance in Cattle in Zimbabwe by 16S rRNA Gene Sequencing. The Open Microbiology Journal 9: 38-42
- Trotha R, Hanck T, Konig W, Konig B (2001) Rapid ribosequencing - an effective diagnostic tool for detecting microbial infection. Infection 29: 12-16
- Henkle E, Winthrop KL (2015) Nontuberculous mycobacteria infections in immunosuppressed hosts.Clinics in Chest Medicine 36: 91-99







