Keywords
E. coli, ESBL, blaTEM, blaSHV, blaCTX-M1, Togo
Background
Infectious diseases are still a major public health issue, especially in Africa where they are responsible for more than 70% of morbidity and mortality [1]. They are dominated by bacterial infections responsible of most of nosocomial and community infections. The β-lactam antibiotic family including penicillins and cephalosporins is more often used to treat these bacterial infections because of their broad-spectrum, low toxicity, efficiency and low cost [2]. Unfortunately, during the last decade, a significant increase in bacterial resistance to these antibiotics was observed. Indeed, according to the World Health Organization (WHO) 2014 report on the global surveillance of antimicrobial resistance in the world, antibiotic resistance has become a serious threat to public health [3]. The emergence and spread of bacteria resistant to β-lactam usually occur by mechanisms of acquisition of exogenous genes of resistance, as plasmid. These genes can inhibit the action of β-lactam antibiotics through production of cephalosporinase (AmpC), Extended Spectrum β-lactamase (ESBL) or carbapenemase, a consequence of the inappropriate use of β-lactam antibiotics in human, animal and in agriculture [4].
Discovered in the early 1980s, ESBLs are a large family of plasmid borne bacterial enzymes belonging to the class A or D of the Ambler classification and found mainly in Enterobacteriaceae [5,6]. Since their discovery and until 1990, most of ESBL detected were TEM and SHV types that disseminated mostly within hospital clones of Klebsiella pneumoniae and Enterobacter spp and associated with nosocomial outbreaks in intensive care unit. These recent years a new type of plasmid encoded β-lactamase CTX-M which spread in Escherichia coli strains and cause urinary tract infections in the communities has been discovered. The blaTEM, blaSHV and blaCTX-M genes, respectively encoded the production of ESBL TEM, SHV and CTX-M, have been described in several epidemiological studies in Europe, Asia, USA, and South America [7-10]. The bacterial species expressing these genes are widespread in the world but some geographical areas have a significantly higher prevalence rates such as South America, Asia and Europe [11].
In Africa, the prevalence of ESBL in Enterobacteriaceae was recently estimated at less than 15% [12], with a predominance of CTX-M-15, and also the AmpC cephalosporinase and carbapenemase [13].
In Togo, an analysis of hospital data on the resistance of blood culture bacteria in the Sylvanus Olympio Teaching Hospital of Lomé conducted in 2009 revealed a prevalence rate of 17.6% of ESBL producing Enterobacteriaceae [14]. However, no information was available about the different types of β- lactamase genes carried by these Enterobacteriaceae.
This study aimed to detect and characterize the resistance genes harbored by ESBL producing E. coli strains isolated from different biological specimens at the National Institute of Hygiene (INH) at Lomé, Togo.
Methods
Collection and identification of E. coli strains, antibiotic susceptibility and detection of ESBL phenotype
The E. coli strains were collected during a descriptive study from May 2013 to July 2015 at the National Institute of Hygiene, Lomé in Togo. INH is the national reference laboratory for the biomedical analysis, epidemiological surveillance, water and food quality control in the country. The strains were isolated from various biological specimen including urine, vaginal samples, semen, urethral curettage (CU), wound swabs and stool. They have been then purified on culture media (Mac- Conkey or Eosin Methylene Blue agar) and identified by API 20E identification system (Biomerieux, Marcy-Etoile, France).
The antibiotic susceptibility testing was performed using the disc diffusion method of Kirby and Bauer [15] on Mueller-Hinton agar (MH) and interpreted using the criteria of the 2014 Antibiogram Committee of the French Society of Microbiology [16]. The antibiotics (Bio-Rad, Marnes-la-Coquette, France) tested were amoxicillin+clavulanic acid (AMC, 20/10 μg), piperacillin+tazobactam (TZP, 75/10 μg), cefoxitin (FOX, 30 μg), ceftriaxon (CRO, 30 μg), ceftazidim (CAZ, 30 μg), cefotaxim (CTX, 30 μg), cefepim (FEP, 30 μg), aztreonam (ATM, 30 μg), imipenem (IMP, 10 μg), amikacin (AN, 30 μg) gentamicin (GM, 15 μg), nalidixic acid (NA, 30 μg) ciprofloxacin (CIP, 5 μg), trimethoprim/ sulfamethoxazole (SXT 1.25/23,75 μg), fosfomycin (FOS, 50 μg), chloramphenicol (C, 30 μg ).
All isolates were subjected to the double disc synergy test for ESBL detection [17]. The presence of ESBL is detected by a synergy between ceftazidim and cefotaxim or ceftriaxon discs and amoxicillin+clavulanic acid disc. Escherichia coli ATCC 25922 was used as control strain for antibiotic susceptibility testing.
E. coli strains resistant to at least one third generation cephalosporin tested, ceftazidim, cefotaxim or ceftriaxon were collected and conserved at -80°C in trypticase soy broth (TCS). They were then sended (in triple packaging boxes with ice packs) to the Biomolecular and Genetic Laboratory (LABIOGENE); Pietro Annigoni Biomolecular Research Center (CERBA), Ouagadougou, Burkina Faso.
Extraction of bacterial DNA
From TCS broth, strains were reactivated on TCS agar for 18-24 h and then inoculated in Luria Bertani (LB, 2 ml). After 18-24 h of overnight culture, LB broth were centrifuged at 10285 g for 10 min and the pellet suspended in 500 μl of phosphate buffer (100 mM, pH 7) to weaken the membranes. The mix was heated at 100°C for 15 min in a water bath to release the genetic material. The DNA is then precipitated in 250 μl of absolute ethanol, washed twice in 1000 μl of ethanol 75%, dried and resuspended in 100 μl of sterile water.
Detection of ESBL genes
The thermocycler GeneAmp PCR System 9700 (Applied Biosystems, California, USA) was used for the amplification of ESBL genes blaTEM, (TEM1, TEM2), blaSHV (SHV1), blaCTX-M-G1 (CTX-M-1, 3 or 15), blaCTX-M-G2 and blaCTX-M-G9 (CTX-M-9 and CTX-M-14). Primers used were purchased from Applied Biosystems (California, USA) (Table 1). Three PCR were performed: two multiplex for TEM/SHV, CTX-M-G2 and CTX-MG9 group [18] and a simplex for CTX-M-G1 [19].
| PCR target |
Sequence (5’–3’) |
Fragment size (pb) |
References |
| TEM |
For : CATTTCCGTGTCGCCCTTATTC |
800 |
[18] |
| Rev : CGTTCATCCATAGTTGCCTGAC |
| SHV |
For : AGCCGCTTGAGCAAATTAAAC |
713 |
[18] |
| Rev : ATCCCGCAGATAAATCACCAC |
| CTX-M-G1 |
For : GTTACAATGTGTGAGAAGCAG |
1000 |
[19] |
| Rev: CCGTTTCCGCTATTACAAAC |
| CTX-M-G2 |
For : CGTTAACGGCACGATGAC |
404 |
[18] |
| Rev : CGATATCGTTGGTGGTRCCA* |
| CTX-M-G9 |
For : TCAAGCCTGCCGATCTGGT |
561 |
[18] |
| Rev : TGATTCTCGCCGCTGAAG |
*R=A ou G
Table 1: Primers used for PCR amplification of bla genes identification.
The PCR were performed in a total volume of 50 μl containing to 2 μl of DNA, 25 μl of 1X AmpliTaq Gold (Master Mix), 5 μl Enhancer, 2 μl of each primer at different concentration (0.2 to 0.4 pmol/μl) and sterile water. The cycling conditions were as follow:
• BlaTEM/SHV and blaCTX-G2-M/G9: initial denaturation 94°C for 10 min. 30 cycles of denaturation 94°C for 40s, annealing 60°C for 40s and extension 72°C for 1 min. Final extension 72°C for 7 min.
• BlaCTX-M-G1 initial denaturation 96°C for 10 min. 35 cycles of denaturation 94°C for 1 min, annealing 50°C for 1 min and extension 72°C for 1 min. Final extension 72°C for 10 min.
• The PCR products were separated on 2% agarose gel staining with ethidium bromide and visualized under UV illumination. A 100 bp DNA ladder (Promega, USA) was used as a marker size.
Statistical analysis
Statistical significance was determined by Fisher's exact test used for small samples size. Analyses were performed using Epi Info Version 7.1.1.14 and a p-value <0.05 was considered to be statistically significant.
Results
Bacterial strains
From May 2013 to July 2015, 91 E. coli strains, resistant at least to one third generation cephalosporin, ceftazidim, cefotaxim, ceftriaxon, were isolated from urine 58/91 (63.74%), vaginal samples 17/91 (17.68%), wound swabs 7/91 (7.69%), 4/91 semen samples (4.40%), urethral curettage (CU) 2/91 (2.20%), sputum 1/91 (1.1%), stool 1/91 (1.1%) and joint fluid 1/91 (1.1%).
Antibiotic susceptibility profile
All E. coli strains were resistant to ceftriaxon and cefotaxim and 98% to ceftazidim. The resistance rate to other β-lactam antibiotics was respectively 2.20%, 17.59% and 25.35% for imipenem (very low levels), piperacillin-tazobactam and cefoxitin. Quinolones, nalidixic acid and ciprofloxacin, trimethoprim/sulfamethoxazole and chloramphenicol were very resistant with rates of 96.67%, 94.50%, 94.44% and 59.09% respectively. Among the aminoglycosides, gentamicin was the most resistant antibiotic (75.82%) in contrast to amikacin (3.30%) which showed also very low levels of resistance. The results are shown in (Table 2).
| ATB |
CAZ |
CRO |
CTX |
FOX |
FEP |
ATM |
TZP |
IPM |
GM |
AN |
NA |
CIP |
C |
FOS |
SXT |
| R (n/N) |
79/91 |
91/91 |
91/91 |
18/71 |
90/91 |
90/91 |
16/91 |
20/91 |
69/91 |
30/91 |
87/90 |
86/91 |
52/88 |
40/91 |
85/90 |
| R (%) |
86.81 |
100 |
100 |
25.35 |
98.9 |
99 |
17.58 |
2.2 |
75.8 |
3.3 |
96.7 |
94.5 |
59.09 |
4.44 |
94.44 |
ATB: antibiotic, R resistant, CAZ ceftazidim, CRO ceftriaxon, CTX cefotaxim, FOX cefoxitin, FEP cefepim, ATM aztreonam, TZP piperacillin-tazobactam, IPM imipenem, GM gentamicin, AN amikacin, NA nalidixic acid, CIP ciprofloxacin, C chloramphenicol, FOS fosfomycin, SXT trimethoprim/sulfamethoxazole.
Table 2: Antibiotic susceptibility profile in ESBL producing E. coli.
In short, resistance to third generation cephalosporins in E. coli has been associated with a resistance to quinolone, trimethoprim/sulfamethoxazole, gentamicin and chloramphenicol. The double disc synergy test was positive in 85/91 strains giving an ESBL presumption rate of 93.40%.
Detection of ESBL genes
PCR was performed to determine the presence of ESBL genes and all E. coli strains, positive or not to the double disc synergy test, carried at least one genes blaTEM, blaSHV and blaCTX-M-G1 These results are shown in (Figure 1). The findings revealed a low presence of ESBL genes alone 19/91 (20.88%) in contrast to percentage of associations, 72/91 (79.21%) E. coli strains harbored more than one ESBL resistance gene. SHV was absent, TEM present in one strain (1.10%) and CTX-M1, in 18/91 (19.78%) strains. The proportion of combinations TEM/SHV, TEM/CTX-M1, were respectively 1/91 (1.10%) and 52/91 (57.14%) and the triple combination TEM/SHV/CTX-M1 was found in 19/91 (20.88%) strains. BlaCTX-M-G2 and blaCTX-M-G9 were not found in our study.
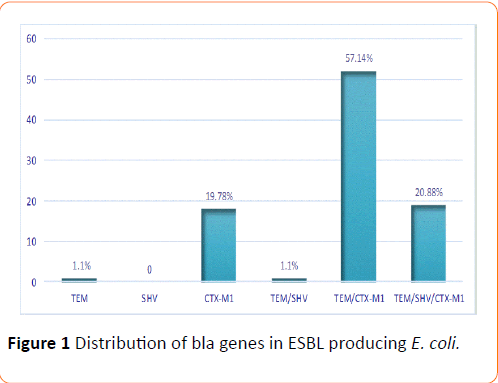
Figure 1: IL-1a profile among categories of seropositive volunteers
Figure 1: Distribution of bla genes in ESBL producing E. coli.
The antibiotic resistance profile in ESBL producing E. coli isolated from urine is similar to that of non-urinary specimens (vaginal samples, wound swabs, semen samples, urethral curettage, sputum, stool, joint fluid) for cefoxitin, imipenem, gentamicin, amikacin, ciprofloxacin, chloramphenicol, and trimethoprim/sulfamethoxazole. The differences of prevalence observed for piperacillin-tazobactam and fosfomycin resistance and for the distribution of ESBL genes were not statistically significant (Table 3).
| Antibiotics % (n/N) |
Urinary specimen 63.74% (58/91) |
Non-Urinary specimen 36.26% (33/91) |
p-value |
| Cefoxitin |
25.35 (12/47) |
25 (6/24) |
1.00 |
| Piperacillin-tazobactam |
22.41 (13/58) |
9.09 (3/33) |
0.154 |
| Imipenem |
1.72 (1/58) |
3.03 (1/33) |
1.00 |
| Gentamicin |
77.59 (45/58) |
72.72 (24/33) |
0.619 |
| Amikacin |
3.45 (2/58) |
3.03 (1/33) |
1.00 |
| Nalidixic acid |
96.55 (56/58) |
96.88 (31/32) |
1.00 |
| Ciprofloxacin |
94.83 (55/58) |
93.94 (31/33) |
1.00 |
| Chloramphenicol |
55.65 (34/57) |
58.06 (18/31) |
1.00 |
| Fosfomicyn |
6.90 (4/58) |
0 (0/32) |
0.293 |
| Trimethoprim/sulfamethoxazole |
94.84 (55/58) |
93.75 (30/32) |
1.00 |
| TEM |
0 |
3.03 (1/33) |
- |
| TEM/SHV |
0 |
3.03 (1/33) |
- |
| CTX-M |
22.41 (13/58) |
15.15 (5/33) |
0.585 |
| TEM/CTX-M |
58.62 (34/58) |
54.55 (18/33) |
0.826 |
| TEM/SHV/CTX-M |
18.97 (11/58) |
24.24 (8/33) |
0.598 |
Table 3: Antibiotic susceptibility profile and bla genes distribution in ESBL producing E. coli from urinary specimen versus non urinary specimen.
Discussion
Enterobacteriaceae are among the most frequently isolated pathogens in biological specimens and involved in bacterial infections with a predominance of E. coli [20]. Antibiotics, β- lactams, aminoglycosides, quinolones, frequently used to cure in these infections are becoming increasingly less and less effective [4]. Indeed, the acquisition of antibiotic resistance factors is due along others, to the excessive and uncontrolled use of antibiotics especially in poor hygiene conditions in developing countries [21,22]. So, the accurate identification of the pathogen and its antibiotic susceptibility profile are necessary for appropriate and effective use of antibiotics and also for a resistance surveillance strategy. The threat of antibiotic resistance in the world is real and each country according to WHO recommendations must develop an antimicrobial resistance surveillance system. In Togo, the prevalence of ESBL producing Enterobacteriaceae was estimated at 17% in laboratory blood cultures [14]. The presence of hospital-acquired ESBL producing Enterobacteriaceae is therefore a reality in Togo and these resistance genes can spread rapidly within the community [23].
In this study, the low frequency of isolation of E. coli resistant at least to a third generation cephalosporin (approximatively 6 strains per month) in a non-hospital laboratory could predict their low prevalence in the community. On the other hand, the prevalence of ESBL phenotype among E. coli resistant at least to one third generation cephalosporin was high at 93.4%. This prevalence is higher than 75% observed in Burkina Faso on E. coli also resistant to third generation cephalosporin [24]. This difference could be explained by an hyperproduction of β- lactamase in strains isolated at Lomé or a poor phenotypic detection due to the limited sensitivity of the double disc synergy test as indicated by Livermore [2].
The presence of ESBL was also associated with a resistance to quinolone, trimethoprim-sulfamethoxazole, gentamicin and chloramphenicol leading these bacteria to become multiresistant. This co-resistance was also observed in a study in Benin, on clinical ESBL producing E. coli from various biological specimens with 100% resistance rate to ciprofloxacin and trimethoprim/sulfamethoxazole and 82.7% for gentamicin [25]. The same trend is observed in other African countries such as Sudan and the Democratic Republic of Congo with resistance rates of 81.4% and 96.5% for ciprofloxacin and 98.6% and 96.5% for trimethoprim/sulfamethoxazole [26,27]. If the resistance to quinolones and trimethoprim/sulfamethoxazole remains generally high above 90%, that of gentamicin is variable with a rate of 48.6% in Sudan [26] and 48.1% in Tanzania on ESBL producing E. coli from clinical urinary specimens [28]. This coresistance, frequently observed in ESBL producing E. coli is due to the existence of other mechanisms of resistance in addition to the β-lactamase gene on the same plasmid, such as the aac(6')- Ib-cr gene which induces resistance to both aminoglycosides and fluoroquinolones [5].
In this study, a decrease in susceptibility to piperacillintazobactam and cefoxitin was observed with the respective resistance rate 17% and 25%. With carbapenems, these antibiotics are among those used for the treatment of infections caused by ESBL producing Enterobacteriaceae including E. coli [29,30]. Resistance to cefoxitin and piperacillin-tazobactam might be explained by the loss or alteration of porin outer membrane or the coexistence of AmpC cephalosporinase with β- lactamase [5]. The resistance of ESBL producing E. coli to cefoxitin and piperacillin-tazobactam is highly variable. On clinical urinary ESBL producing E. coli strains resistance to piperacillin-tazobactam in India, China and Japan was respectively 10.1%, 6.3% and 13.7% [31-33]. Resistance to cefoxitin was 41% in Cambodia [34], and 0% in Morocco [35]. The presence of this co-resistance could limit the choice of antibiotics to be used in case of ESBL producing E. coli infection. Imipenem and amikacin, with a low resistance rate at 2.20% and 3.30% respectively remained the antibiotics of choice for nonurinary infection with ESBL producing strains [30,36]. Fosfomycin, with a resistance rate at 4.40% for ESBL producing E. coli still remains one of the efficient treatment in case of urinary tract infection [37] caused by E. coli [38]. Nevertheless, the emergence of resistance to these antibiotics, advocates the establishment of a more enhanced surveillance strategy.
For the presence of resistance genes, PCR was positive for blaTEM, blaSHV and blaCTX-M-G1 in all E. coli strains and negative for blaCTX-M-G2 and blaTX-M-G9 genes. These results showed that the combination TEM/CTX-M1 (57%) dominated the resistance profile in ESBL producing E. coli followed by the combinations TEM/SHV/CTX-M1 (20.8%) and CTX-M1 (19.7%). The blaTEM gene and the combination blaTEM/SHV were less frequent with a prevalence of 1.1% each. The worldwide expansion of the community acquired ESBL gene, CTX-M [39] is thus confirmed in this study and also its association with TEM and SHV genes, previously reported in studies on E. coli from urinary specimens in Europe [40], Asia [41,42], America [43,44] and North Africa [35].
In West Africa, recent studies have also reported the prevalence of CTX-M gene and its association with the TEM and SHV in ESBL producing E. coli and isolated from various biological specimens. Thus in Burkina Faso, Zongo et al. had reported on 96 the following rates of these genes: TEM 25/96, SHV 20/96, CTX-M 70/96 and TEM/SHV/CTX-M 6/96 [24]. In Senegal, Dia et al., found on 32 E. coli: TEM 9/32, SHV 1/32, CTX-M15 29/32 and CTX-M15/TEM 13/32 [45]. In Benin, Anago et al. had found 74.2% for TEM, 24.1% for SHV and 17.4% for TEM/SHV combination, CTX-M was not determined [25]. The combination of ESBL genes is more common in this study 72/91, 79.12% than in Burkina Faso, with 6/96, 6.96% [24], in Senegal, 13/32, 40.63% [45] and in Morocco, 2/7, 28.57% [35].
It is especially noted that the resistance genes are most often in combination (79.12% of associated genes versus 20.88% of genes alone). The absence of the first genes BLSE (TEM and SHV) solo as opposed to the new CTX-M-G1 gene found alone in 18 strains suggests a progressive consolidation of resistance genes on a single mobile genetic element (plasmids, Integron, etc.)
The CTX-M-G1 includes CTX-M-1, CTX-M-3 or CTX-M-15 types. The CTX-M β-lactamase belong to the class A of β-lactamase enzymes described for the first time in Western Europe and therefore found worldwide [6,46]. The CTX-M-15 type that differs from CTX-M-3 by Asp240Gly substitution conferring a resistance to ceftazidim [5,46] is the most widespread [46]. In the absence of more detailed results on the characterization of CTX-M1 gene and face to a high level of resistance to ceftazidim (100%) in this study, we can suppose that they are likely to carry the CTX-M15 gene. The prevalence of this gene described in some studies in West Africa, Senegal [45,47], Niger [48], Nigeria [49-51] would suggest its wide distribution in that part of Africa.
In this study, the prevalence of ESBL genes and the different antibiotic susceptibility profiles distribution is independent of biological specimen (urinary specimen versus non urinary specimen) in which ESBL producing E. coli was isolated. The small sample size could explain this finding.
Conclusions
This study showed that E. coli strains isolated at the National Institute of Hygiene in Lomé and resistant to third generation cephalosporins generally produce extended spectrum betalactamase (ESBL). The results revealed the high prevalence of CTX-M-G1 alone or in combination with TEM and SHV genes. ESBL production in these strains was associated with a resistance to quinolones, trimethoprim/sulfamethoxazole, gentamicin and chloramphenicol. But they remain largely susceptible to imipenem, amikacin and fosfomycin allowing an alternative therapy for infections by ESBL producing E. coli strain.
Antibiotic resistance by ESBL production is a reality worldwide and particularly in Togo. Their wide dissemination jeopardize the treatment of common infections in the community and in hospitals. The need of molecular characterization of these resistance mechanism to antibiotics is necessary for better therapeutic management of infections and also to contribute to the establishment of an antibiotic resistance surveillance system.
This is a first determination of the β-lactamase genes in Togo.
Declarations
Ethical approval
This study has received an approval for the transfer of E. coli strains from INH Lomé, Togo to the Biomolecular and Genetic Laboratory (Labiogene); Pietro Annigoni Biomolecular Research Center (Cerba), University of Ouaga I, Prof. Joseph Ki Zerbo, Burkina Faso and have been accepted by the institutional review board of CERBA/LABIOGENE.
Availability of data
The database analyzed during the study is available on reasonable request from the corresponding author.
Author's contribution
FS, SD and JS designed the study, FS, AS, SS, AO, TZ and AD performed the experiments, FS, CN and SD analyzed the data and wrote the manuscript which was approved by all the other co-authors.
Acknowledgement
We thank Romaric Bado for his assistance in statistical analysis. We are grateful to the Department of Bacteriology personnel at INH at Lomé, Togo, for their help during the collection of E. coli strains.
Funding
FS was granted from the Islamic Development Bank for her master degree at the University of Ouaga I, Prof. Joseph Ki Zerbo, Burkina Faso. This work has been supported by West African Economic and Monetary Union (WAEMU) Commission, Ouagadougou, Burkina Faso.
Conflict of Interest
The authors declare no conflict of interest.
17996
References
- Statistiques Sanitaires Mondiales 2012 (2012) World Health Organization.
- Livermore DM (1995) Beta-Lactamases in laboratory and clinical resistance. Clin Microbiol Rev 8:557-84.
- World Health Organization (2014) Antimicrobial resistance: global report on surveillance. Geneva, Switzerland: World Health Organization.
- Meyer E, Gastmeier P, Deja M, Schwab F (2013) Antibiotic consumption and resistance: Data from Europe and Germany. Int J Med Microbiol 303:388-395.
- Bradford PA (2001) Extended-Spectrum β-Lactamases in the 21st Century: Characterization, Epidemiology, and Detection of This Important Resistance Threat. Clin Microbiol Rev 14:933-951.
- Paterson DL, Bonomo RA (2005) Extended-Spectrum β-Lactamases: a Clinical Update. Clin Microbiol Rev 18:657-686.
- Bush K(2008) Extended-spectrum β-lactamases in North America, 1987-2006. Clin Microbiol Infect 14:134-143.
- Cantón R, Novais A, Valverde A, Machado E, Peixe L, et al. (2008) Prevalence and spread of extended-spectrum β-lactamase-producing Enterobacteriaceae in Europe. Clin Microbiol Infect 14:144-153.
- Hawkey PM (2008) Prevalence and clonality of extended-spectrum β-lactamases in Asia. Clin Microbiol Infect 14:159-165.
- Villegas MV, Kattan JN, Quinteros MG, Casellas JM (2008) Prevalence of extended-spectrum β-lactamases in South America. Clin Microbiol Infect 14:154-158.
- Dhillon RH, Clark J (2012) ESBLs: A Clear and Present Danger? Crit Care Res Pract.
- Tansarli GS, Poulikakos P, Kapaskelis A, Falagas ME (2014) Proportion of extended-spectrum β-lactamase (ESBL)-producing isolates among Enterobacteriaceae in Africa: evaluation of the evidence-systematic review. J Antimicrob Chemother 69:1177-1184.
- Storberg V (2014) ESBL-producing Enterobacteriaceae in Africa-a non-systematic literature review of research published 2008-2012. Infect Ecol Epidemiol 4: 20342
- Salou M, Assimadzi K, Wateba IM, Dossim S, Tigossou SD, et al. (2011) Résistance aux antibiotiques des bactéries isolées en 2009 au laboratoire de bactériologie du Chu-Tokoin Lome-Togo. J Rech Sci Univ Lome13:151-159.
- Bauer AW, Kirby WMM, Sherris JC, Turck M (1999) Antibiotic susceptibility testing by a standardized single disk method. Microbiol Centen Perspect 40-45.
- Bonnet R, Bru JP, Caron F, Cattoir V, Chardon H, et al. (2014) Comité de l’Antibiogramme de la Société Française de Microbiologie/EUropean Committee on Antimicrobial Susceptilility Testing: Recommandations.
- Jarlier V, Nicolas MH, Fournier G, Philippon A (1988) Extended broad-spectrum beta-lactamases conferring transferable resistance to newer beta-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev Infect Dis 10:867-878.
- Dallenne C, Da Costa A, Decre D, Favier C, Arlet G (2010) Development of a set of multiplex PCR assays for the detection of genes encoding important β-lactamases in Enterobacteriaceae. J Antimicrob Chemother 65:490-495.
- Pagani L, Dell’Amico E, Migliavacca R, D’Andrea MM, Giacobone E, et al. (2003) Multiple CTX-M-Type Extended-Spectrum β-Lactamases in Nosocomial Isolates of Enterobacteriaceae from a Hospital in Northern Italy. J Clin Microbiol 41:4264-4269.
- Zhanel GG, DeCorby M, Adam H, Mulvey MR, McCracken M, et al.(2010) Prevalence of Antimicrobial-Resistant Pathogens in Canadian Hospitals: Results of the Canadian Ward Surveillance Study (CANWARD 2008). Antimicrob Agents Chemother 54:4684-4693.
- Okeke IN, Lamikanra A, Edelman R (1999) Socioeconomic and behavioral factors leading to acquired bacterial resistance to antibiotics in developing countries. Emerg Infect Dis 5:18-27.
- Kariuki S, Dougan G (2014) Antibacterial resistance in sub-Saharan Africa: an underestimated emergency: Antibacterial resistance in sub-Saharan Africa. Ann N YAcad Sci 1323:43-55.
- Tande D, Boisrame-Gastrin S, Munck MR, Hery-Arnaud G, Gouriou S, et al. (2010) Intrafamilial transmission of extended-spectrum-β-lactamase-producing Escherichia coli and Salmonella enterica Babelsberg among the families of internationally adopted children. J Antimicrob Chemother 65:859-865.
- Zongo KJ, Metuor Dabire A, Compaore LG, Sanou I, Sangare L (2015) First detection of bla TEM, SHV and CTX-M among Gram negative bacilli exhibiting extended spectrum beta-lactamase phenotype isolated at University Hospital Center, Yalgado Ouedraogo, Ouagadougou, Burkina Faso. Afr J Biotechnol 14:1174-1180.
- Anago E, Ayi-Fanou L, Akpovi CD, Hounkpe WB, Agassounon-Djikpo Tchibozo M, et al. (2015) Antibiotic resistance and genotype of beta-lactamase producing Escherichia coli in nosocomial infections in Cotonou, Benin. Ann Clin Microbiol Antimicrob 14:5.
- Ibrahim ME, Bilal NE, Magzoub MA, Hamid ME (2013) Prevalence of Extended-spectrum β-Lactamases-producing Escherichia coli from Hospitals in Khartoum State, Sudan. Oman Med J 28:116-120.
- Irenge LM, Kabego L, Vandenberg O, Chirimwami RB, Gala JL (2014) Antimicrobial resistance in urinary isolates from inpatients and outpatients at a tertiary care hospital in South-Kivu Province (Democratic Republic of Congo). BMC Res Notes 7:374.
- Moyo SJ, Aboud S, Kasubi M, Lyamuya EF, Maselle SY (2010) Antimicrobial resistance among producers and non-producers of extended spectrum beta-lactamases in urinary isolates at a tertiary Hospital in Tanzania. BMC Res Notes 3:348.
- Peterson LR (2008) Antibiotic policy and prescribing strategies for therapy of extended-spectrum β-lactamase-producing Enterobacteriaceae: the role of piperacillin-tazobactam. Clin Microbiol Infect 14:181-184.
- Kanj SS, Kanafani ZA (2011) Current Concepts in Antimicrobial Therapy Against Resistant Gram-Negative Organisms: Extended-Spectrum β-Lactamase-Producing Enterobacteriaceae, Carbapenem-Resistant Enterobacteriaceae, and Multidrug-Resistant Pseudomonas aeruginosa. Mayo Clin Proc 86:250-259.
- Gupta V, Rani H, Singla N, Kaistha N, Chander J (2013) Determination of extended-spectrum β-lactamases and ampc production in uropathogenic isolates of Escherichia coli and susceptibility to fosfomycin. J Lab Physicians 5:90.
- Hu YY, Cai JC, Zhou HW, Chi D, Zhang XF, et al. (2013) Molecular Typing of CTX-M-Producing Escherichia coli Isolates from Environmental Water, Swine Feces, Specimens from Healthy Humans, and Human Patients. Appl Environ Microbiol 79:5988-5996.
- Asakura T, Ikeda M, Nakamura A, Kodera S (2014) Efficacy of empirical therapy with non-carbapenems for urinary tract infections with extended-spectrum beta-lactamase-producing Enterobacteriaceae. Int J Infect Dis 29:91-95.
- Ruppé E, Hem S, Lath S, Gautier V, Ariey F, et al. (2009) CTX-M β-Lactamases in Escherichia coli from Community-acquired Urinary Tract Infections, Cambodia. Emerg Infect Dis 15:741-748.
- Bourjilat F, Bouchrif B, Dersi N, Claude JD, Amarouch H (2011) Emergence of extended-spectrum beta-lactamases-producing Escherichia coli in community-acquired urinary infections in Casablanca, Morocco. J Infect Dev Ctries 5:850-855.
- Curello J, MacDougall C (2014) Beyond Susceptible and Resistant, Part II: Treatment of Infections Due to Gram-Negative Organisms Producing Extended-Spectrum β-Lactamases. JPediatr Pharmacol Ther 19:156-164.
- Gupta K, Hooton TM, Naber KG, Wullt B, Colgan R, et al. (2011) International Clinical Practice Guidelines for the Treatment of Acute Uncomplicated Cystitis and Pyelonephritis in Women: A 2010 Update by the Infectious Diseases Society of America and the European Society for Microbiology and Infectious Diseases. Clin Infect Dis 52:e103-e120.
- Sastry S, Clarke LG, Alrowais H, Querry AM, Shutt KA (2015) Clinical Appraisal of Fosfomycin in the Era of Antimicrobial Resistance. Antimicrob Agents Chemother 59:7355-7361.
- Turner PJ (2005) Extended-Spectrum β-Lactamases. Clin Infect Dis 41:S273-S275.
- van der Donk CFM, van de Bovenkamp JHB, De Brauwer EIGB, De MP, Feldhoff KH, et al. (2012) Antimicrobial Resistance and Spread of Multi Drug Resistant Escherichia coli Isolates Collected from Nine Urology Services in the Euregion Meuse-Rhine. PLoS ONE 7:e47707.
- Zhang J, Zheng B, Zhao L, Wei Z, Ji J, et al. (2014) Nationwide high prevalence of CTX-M and an increase of CTX-M-55 in Escherichia coli isolated from patients with community-onset infections in Chinese county hospitals. BMC Infect Dis 14:1.
- Liu H, Wang Y, Wang G, Xing Q, Shao L, et al. (2015) The prevalence of Escherichia coli strains with extended spectrum beta-lactamases isolated in China. Front Microbiol 6:335.
- Hayakawa K, Gattu S, Marchaim D, Bhargava A, Palla M, et al. (2013) Epidemiology and Risk Factors for Isolation of Escherichia coli Producing CTX-M-Type Extended-Spectrum β-Lactamase in a Large U.S. Medical Center. Antimicrob Agents Chemother 57:4010.
- Bartoloni A, Sennati S, Di Maggio T, Mantella A, Riccobono E, et al. (2016) Antimicrobial susceptibility and emerging resistance determinants (blaCTX-M, rmtB, fosA3) in clinical isolates from urinary tract infections in the Bolivian Chaco Int. J Infect Dis 43:1-6.
- Dia ML, Ngom B, Diagne R, Ka R, Lo S, et al. (2016) Molecular detection of CTX-M-15-type β-lactamases in Escherichia coli strains from Senegal. New Microbes New Infect 9:45-46.
- Bonnet R (2004) Growing Group of Extended-Spectrum β-Lactamases: the CTX-M Enzymes. Antimicrob Agents Chemother 48:1-14.
- Ruppe E, Woerther PL, Diop A, Sene AM, Da Costa A, et al. (2009) Carriage of CTX-M-15-Producing Escherichia coli Isolates among Children Living in a Remote Village in Senegal. Antimicrob Agents Chemother 53:3135-3137.
- Woerther PL, Angebault C, Jacquier H, Hugede HC, Janssens AC, et al. (2011) Massive Increase, Spread, and Exchange of Extended Spectrum β-Lactamase-Encoding Genes Among Intestinal Enterobacteriaceae in Hospitalized Children With Severe Acute Malnutrition in Niger. Clin Infect Dis 53:677-685.
- Iroha IR, Esimone CO, Neumann S, Marlinghaus L, Korte M, et al. (2012) First description of Escherichia coli producing CTX-M-15-extended spectrum beta lactamase (ESBL) in out-patients from south eastern Nigeria. Ann Clin Microbiol Antimicrob 11:19.
- Inwezerua C, Mendonça N, Calhau V, Domingues S, Adeleke OE (2014) Occurrence of extended-spectrum beta-lactamases in human and bovine isolates of Escherichia coli from Oyo state, Nigeria J Infect Dev Ctries8:774-779.
- Raji MA, Jamal W, Ojemeh O, Rotimi VO (2015) Sequence analysis of genes mediating extended-spectrum beta-lactamase (ESBL) production in isolates of Enterobacteriaceae in a Lagos Teaching Hospital, Nigeria. BMC Infect Dis 15:259.






