Research Article - () Volume 11, Issue 1
Gene Polymorphism of Superoxide Dismutase in Gestational Diabetes Mellitus in Pregnant Women
Mai M. Madkour1*,
Abd El-Aziz F. Abd El-Aziz1,
Fardous F. El-Senduny1,
Afaf M. Elsaid2 and
Abd El-Aziz A. El-Refaey3
1Department of Chemistry, Mansoura University, Mansoura, Egypt
2Department of Pediatrics, Mansoura University, Mansoura, Egypt
3Department of Obstetrics and Gynecology, Mansoura University, Mansoura, Egypt
*Correspondence:
Mai M. Madkour,
Department of Chemistry, Mansoura University, Mansoura,
Egypt,
Tel: 01066907311,
Email:
Received: 31-Jul-2022, Manuscript No. IPACLR-22-12895;
Editor assigned: 03-Aug-2022, Pre QC No. IPACLR-22-12895 (PQ);
Reviewed: 18-Aug-2022, QC No. IPACLR-22-12895;
Revised: 23-Dec-2022, Manuscript No. IPACLR-22-12895 (R);
Published:
02-Jan-2023, DOI: 10.36648/2386-5180.22.11.450
Abstract
Background: Gestational diabetes mellitus is the most common metabolic disorder that develops during pregnancy. GDM can cause major health problems for the mother both during pregnancy and after the baby is born. In addition, the metabolism of the mother's kids is abnormal. The study aimed to investigate superoxide dismutase 1 genetic variants in pregnant women diagnosed with gestational diabetes using amplification refractory mutation system analysis.
Results: Our study demonstrated that the frequencies of genotypes in women with GDM were SOD1-AG (90.0%). The study revealed there is a significant difference in the frequency of the genotype of SOD1 between GDM and healthy pregnant women.
Conclusions: To the best of our knowledge, this is the first study to indicate that the genotype of SOD1 (A/G rs2070424) has a significant impact on the development of gestational diabetes mellitus in Egyptian women.
Keywords
Gestational diabetes; Superoxide dismutase 1
gene; Genetic polymorphism; Oxidative stress
Abbreviations
GDM: Gestational Diabetes; SOD1:
Superoxide Dismutase 1; ROS: Reactive Oxygen Species;
SNPs: Single-Nucleotide Polymorphisms; T-ARMS-PCR: Tetra-
Primer Amplification Refractory Mutation System
INTRODUCTION
Gestational Diabetes Mellitus (GDM) is defined as any degree
of glucose intolerance that develops or is detected during
pregnancy. In the second or third trimester, pregnant women are
diagnosed [1]. This condition is associated with adverse
pregnancy outcomes, including fetal macrosomia, stillbirth, neonatal metabolic disturbances, and related problems. GDM is
not caused by a lack of insulin but by other hormones produced
during pregnancy that can make insulin less effective, a
condition referred to as insulin resistance [2].
Oxidative stress is a general term used to describe the steady
state of oxidative damage in a cell, tissue, or organ caused by
Reactive Oxygen Species (ROS). The majority of ROS is produced
endogenously as a byproduct of normal and necessary
reactions, such as energy generation from mitochondria. This
oxidative stress was found to be greater in women with GDM
than in normal pregnant women [3]. A complex and integrated
antioxidant system plays a crucial role in protecting cells or
tissues from damage as a result of ROS. The expression and
activity of antioxidants are changed during oxidative stress [4].
Human Cu–Zn Superoxide Dismutase (SOD1) gene is located
on chromosome 21 (21q22.1), a homo-dimeric metalloprotein
consisting of 153 amino acids with the two subunits linked noncovalently
[5]. The SOD1 polymorphism (rs2070424) is
distinguished by the substitution of Adenine (A) to Guanine (G)
at codon 251 in intron 3 of the gene. This polymorphism has
been linked to a reduction in antioxidant capacity [6]. Actually,
the SOD1 gene encodes an enzyme that catalyzes the
dismutation of superoxide into O2 and H2O2 to eliminate free
superoxide radicals produced in the body. The delicate balance
between the elimination of ROS and limiting oxidative stress is
disrupted by genotype related differences in enzyme activity.
The study aimed to evaluate the Single Nucleotide
Polymorphisms (SNPs) in SOD1 (A/G rs2070424) and correlate its
genotyping to gestational diabetes mellitus. To the best of our
knowledge, this is the irst study to report the association of
their genotypes with gestational diabetes in Egyptian women.
Materials and Methods
This case-control study was conducted with 70 women with
gestational diabetes and 70 healthy pregnant women. They
were recruited from the obstetrics and gynecology department, faculty of medicine, Mansoura university in the period from July
2017 to March 2019. All of the women in the study were at least
18 years old and had been diagnosed at a gestational age of
more than 24 weeks.
Pregnant women with type 1 diabetes, previous macrosomia
(a baby weighing more than 4000 g at birth), polycystic ovary
syndrome, and other serious medical conditions (hypertension,
renal disease, moderate to severe anaemia, thyroid disorder)
that interfered with maternal and perinatal outcomes were
excluded.
Sample collection and DNA extraction
Each patient's fasting blood was drawn and dispensed into an
EDTA containing tube. A blood sample was divided into two
portions (3 mL and 2 mL). After centrifugation at >2000 g for 10
minutes, the first portion was used to collect plasma. The
plasma was extracted and used for biochemical analysis as well
as a native-PAGE investigation of SOD activity. The second
portion was used to extract DNA. The DNA quantity and quality
were measured by reading the absorbance at λ 230 nm and λ
260 nm.
Amplification refractory mutation system (ARMS)-
PCR analysis Tri-ARMS-PCR analysis of superoxide
dismutase 1 gene (SOD1)
Primers used in this study (F1, F2 and R) are provided in Table
1. In the SOD1 gene (A/G rs2070424).
| Primer |
Sequence (5′‐3′) |
Fragment size |
| F1 |
TAGCTTTGTTAGCTATGCCA |
A allele: 480 (F1+R) |
| F2 |
TAGCTTTGT TAGCTATGCCG |
G allele: 480 (F2+R) |
| R |
ATCTTTAGAA ACCGCGACTA |
|
| F1=Forward primer 1, F2=Forward primer 2, R=Reverse primer |
Table 1: Primers used in Tri‐ARMS‐PCR of SOD1 gene (A/G rs2070424).
200 ng of genomic DNA, 3 μL of an allele primers (F1 and RP)
or G allele primers (F2 and RP) (Table 1) and 8 μL 2 × Taq Master
Mix (EmeraldAmp GT PCR Master Mix, Cat. No.RR310A) were
used in an 18 μL polymerase chain reaction (PCR). The PCR
protocol was as follows: 95°C for 3 min, then 35 cycles at 95°C
for 30 sec, annealing at 60°C for 30 sec, extension at 72°C for 30
sec, and a final extension at 72°C for 10 min. The agarose gel
electrophoresis (2%) was performed at 125 V and a 100 bp
MaestroGen DNA ladder (Cat. No. 02001_500) was used to
investigate and confirm the quality of the PCR products (Cleaver
Scientific Ltd., UK).
Biochemical measurements
Antioxidant levels were estimated by determining catalase,
GR activity and plasma levels of MDA using kit supplied by
biodiagnostic (Cairo, Egypt). Plasma levels of catalase,
superoxide dismutase and glutathione reductase activity can be
determined according to the methods of Fossati, and Ponti, et
al. and goldberg and spooner respectively [7-10]. Satoh and
Tsikas techniques can be used to detect plasma
malondialdehyde and nitric oxide levels respectively [11].
SOD activity gel method
The SOD activity gel test was developed by a method that is
based on the suppression of NBT reduction by SOD. This test is
based on the ability of O2•−to interact with NBT and convert the
tetrazolium (yellow colour) in the gel to a blue aggregate. SOD
active areas generate a clear region (achromatic bands) that
competes with NBT for O2•− [12-14].
Statistical analysis
Statistical analysis was done using the software package, SPSS
version 22 and excel. data were expressed as mean ± SD. The significance of the difference between women with gestational
diabetes and healthy pregnant women was determined using
ANOVA. Take into account that P–values of less than 0.05 were
statistically significant. Frequencies of either genotype or allele
of SOD1 polymorphism between two groups were analyzed by
the Fisher exact test and hardy Weinberg equilibrium.
Results
Plasma catalase, superoxide dismutase and glutathione
reductase levels were significantly decreased in women with
gestational diabetes when compared with the control group. On
the other hand, the levels of malondialdehyde and nitric oxide
were significantly higher than normal pregnant women (Table 2).
| Parameters |
GDM group (n =70) |
Control group (n =70) |
OR (95% CI) |
P value |
| CAT (U/L) mean ± SD |
337.39 ± 133.06 |
875.54 ± 100.6 |
0.65 (undefined) |
p<0.001* |
| SOD (U/mL) mean ± SD |
1.43 ± 0.38 |
2.32 ± 0.14 |
undefined |
p<0.001* |
| GR (U/L) median (IQR) |
6.43 (4.02-10.45) |
30.54 (22.51-40.21) |
0.54 (0.44-0.65) |
p<0.001* |
| MDA (nmol/mL) mean ± SD |
13.35 ± 4.30 |
5.36 ± 2.83 |
2.15(1.77-2.59) |
p<0.001* |
| NO (mg/L) median (IQR) |
4.81 (3.53-6.79) |
2.04 (1.77-2.78) |
undefined |
P<0.001* |
Table 2: Comparison of antioxidant enzymes and oxidative stress markers for gestational diabetes mellitus in pregnant women with healthy pregnant women.
Genetic polymorphism and genotype frequencies
Analysis of SOD1 gene: The genetic polymorphism in
the SOD1 gene (A251G) was investigated, and the genotypes
were shown in Figure 1. The frequencies of the allele and
genotype of the SOD1 gene (rs2070424) between women
with gestational diabetes and healthy pregnant women were
listed in Tables 3 and 4. Genotype frequencies of the
rs2070424 A/G polymorphism differed significantly
between women with gestational diabetes and control (p=0.025), while allele frequencies did not show any signi icant
difference between women with gestational diabetes and
control. Moreover, the distribution of genotypes of SOD1 for
GDM and control groups was in alignment with the Hardy-
Weinberg equilibrium (χ2=44.84, p<0.001; χ2=70, p<0.001),
which was analyzed by Fisher’s exact test (Table 5).
| Allele |
GDM group |
Control group |
P value |
| |
(n=70) |
(n=70) |
|
| A |
71 (50.7%) |
70 (50%) |
0.905 |
| G |
69 (49.3%) |
70 (50%) |
|
| The data was expressed as frequency and percentage. GDM: gestational diabetes mellitus. |
Table 3: Allele frequencies of SOD1 gene (rs2070424) in the current study.
| Genotype |
GDM group (n=70) |
Control group (n=70) |
P value |
| A/A |
4 (5.7%) |
0 |
|
| A/G |
63 (90.0%) |
70 (100%) |
0.025* |
| G/G |
3 (4.3%) |
0 |
|
| The data was expressed as frequency and percentage. GDM: gestational diabetes mellitus. |
Table 4: Genotype frequencies of SOD1 gene (rs2070424) in the current study.
| |
GDM group (n=70) |
Control group (n=70) |
| HWE |
χ2=44.84 |
χ2= 70 |
| P -Value |
p<0.001* |
p<0.001* |
| GDM: Gestational Diabetes Mellitus; χ2: chi-Square Test; HWE: Hardy-Weignberg Equilibrium; *statistically significant if p<0.05. |
Table 5: Fisher exact test for Hardy-Weinberg equilibrium in the current study.
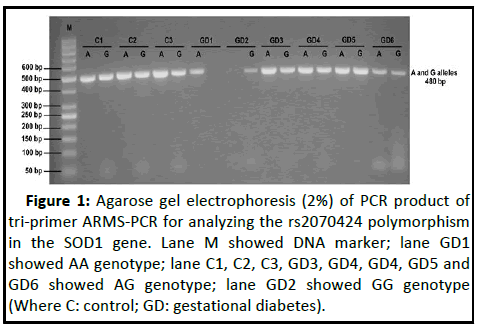
Figure 1: Agarose gel electrophoresis (2%) of PCR product of
tri-primer ARMS-PCR for analyzing the rs2070424 polymorphism
in the SOD1 gene. Lane M showed DNA marker; lane GD1
showed AA genotype; lane C1, C2, C3, GD3, GD4, GD4, GD5 and
GD6 showed AG genotype; lane GD2 showed GG genotype
(Where C: control; GD: gestational diabetes).
The major risk of GDM was evaluated by the co-dominant,
dominant, recessive, and over dominant models as shown in Table 6. In the co-dominant model (A/A vs. A/G vs. G/G), there
was a statistically significant difference between GDM and
healthy pregnant women (p=0.025). The dominant model (A/A
vs. A/G+G/G) showed a significant risk of GDM when compared
with the A/G+G/G genotype (p=0.04). The recessive model (A/A +A/G vs. G/G) did not display any significant risk of GDM
compared to the A/A+A/G genotype (p=0.08). Furthermore, the
over dominant (A/A+G/G vs. A/G) showed a significant risk of
GDM compared to the A/G genotype (p=0.006).
| Model |
Genotype |
GDM group (n=70) |
Control group (n=70) |
OR (95% CI) |
P value |
| Co-dominant |
A/A |
0 |
4 (5.7%) |
undefined |
0.025* |
| A/G |
70 (100%) |
63 (90.0%) |
| G/G |
0 |
3 (4.3%) |
| Dominant |
A/A |
0 |
4 (5.7%) |
undefined |
0.04* |
| A/G+G/G |
70 (100%) |
66 (94.3%) |
| Recessive |
A/A+A/G |
70 (100%) |
67 (95.7%) |
undefined |
0.08 |
| G/G |
0 |
3 (4.3%) |
| Over dominant |
A/A+G/G |
0 |
7 (10%) |
undefined |
0.006* |
| A/G |
70 (100%) |
63 (90%) |
| The data was presented as percentage and frequency. GDM: Gestational Diabetes Mellitus; OR: Odds Ratio; 95% CI: 95% Confidence Interval for the difference in means between the two groups; P is significant when <0.005. |
Table 6: Association between genotypes of SOD1 and response status in the current study.
Correlation between antioxidant enzymes, oxidative
stress markers and SOD1 (rs2070424) SNP
The association between antioxidant enzymes, oxidative
stress markers and the SOD1 gene (rs2070424) SNP in the
population under study was presented in Table 7. The study
population (GDM and control groups) was divided into three
groups according to genotypes in AA, AG, and GG. There wasn’t any signficant difference in the biochemical parameters
between the three groups, except that the level of MDA, and
nitric oxide showed signi icant differences between AG and
GG (p=0.039, p=0.014) respectively.
| Parameters |
AA |
AG |
GG |
Test of significance |
Significance within groups |
| Catalase mean ± SD |
409.63 ± 151.27 |
331.11 ± 134.65 |
372.91 ± 28.94 |
F=0.761 P=0.471 |
P1=0.598 P2=0.720 P3=0.258 |
| Superoxide dismutase mean ± SD |
1.41 ± 0.61 |
1.44 ± 0.38 |
1.23 ± 0.06 |
F=0.391 P=0.678 |
P1=0.383 P2=0.567 P3=0.879 |
| Glutathione reductase Median (IQR) |
10.05 (2.61-17.48) |
6.43 (4.02-10.45) |
4.02 (4.01-13.66) |
KW P=0.754 |
P1=0.951 P2=0.721 P3=0.449 |
| Malondialdehyde Median (IQR) |
11.26 (7.96-17.69) |
12.08 (10.49-17.14) |
9.66 (7.91-9.71) |
KW P=0.109 |
P1=0.039* P2=0.289 P3=0.578 |
| Nitric oxide Median (IQR) |
3.79 (3.41-4.40) |
5.17 (3.75-7.28) |
3.29 (2.82-3.52) |
KW P=0.015* |
P1=0.014* P2=0.108 P3=0.109 |
| F: one way ANOVA test; KW: Kruskal Wallis test; IQR: Interquartile Range; P1: Difference between AG and GG; P2: difference between AA and GG; P3: difference between AA and AG; *statistically significant if p<0.05. |
Table 7: Correlation between antioxidant enzymes, oxidative stress markers and SOD1 (rs2070424) SNP in the current study.
SOD native gel
The native PAGE gel assay facilitated the identification of SOD
isoenzymes in normal pregnant women and women with
gestational diabetes. Figure 2 showed superoxide dismutase
isoenzymes (Mn-SOD and CuZn-SOD) activities in native gel. The
stain intensity of the bands in the control lane was higher,
whereas the stain intensity of the bands in the gestational
diabetes group lanes was lower. Therefore, the activity of SOD
isoenzymes was decreased in women with gestational diabetes
compared to normal pregnant women.
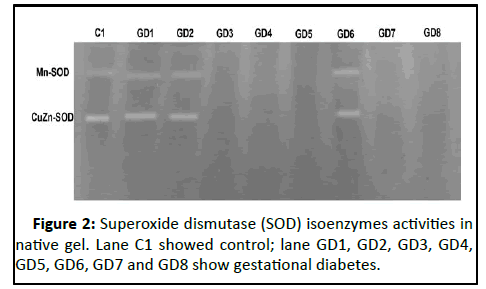
Figure 2: Superoxide dismutase (SOD) isoenzymes activities in
native gel. Lane C1 showed control; lane GD1, GD2, GD3, GD4,
GD5, GD6, GD7 and GD8 show gestational diabetes.
Discussion
Gestational diabetes mellitus is the most common illness
among pregnant women all over the world. Insulin resistance
distinguishes it as a complicated metabolic condition [15]. The
study aimed to find whether there was a link between
superoxide dismutase 1 A251G SNP (rs2070424) with the
incidence of gestational diabetes in Egyptian women.
The antioxidant defense system includes the enzymes CAT,
SOD, and GR. They are in charge of regulating the level of free radicals in cells [16]. In this study, MDA and nitric oxide levels
increased, whereas CAT, SOD, and GR activity decreased in
women with GDM compared to normal pregnant women.
It was discovered that there was a substantial rise in MDA
levels in other studies [17]. MDA levels rise and anti-oxidant
enzyme levels fall as pregnancy progresses [18]. The GDM group
had higher MDA concentrations throughout the pregnancy, with
significant differences in the first and second trimesters [19].
When comparing women with GDM to controls, a significant
increase in lipid peroxidation was found [20].
Previous studies have reported conflicting results related to
NO levels in pregnancy. It was found that NO levels dropped
during pregnancy, whereas NO levels remained constant
throughout the pregnancy in other studies [21-23].
It was discovered that SOD activity was found to be
significantly lower in pregnant females with gestational diabetes
as compared to normal pregnant females [24]. Furthermore,
superoxide dismutase activity is lower in pregnant women than
in non-pregnant women in the third trimester of normal
pregnancy [25,26]. The reduction in SOD levels was
demonstrated by another study [27].
Another research revealed that CAT activity did not alter
throughout pregnancy, despite the fact that CAT activity is
greater during pregnancy [28,29]. Reduced CAT activity might be
attributed to lower lipid peroxidation rates in the third trimester
of pregnancy. In agreement with another study, blood catalase
activity in gestational diabetes is decreased [30]. The decrease in
CAT as an inducible enzyme may be due to the lower level of
H2O2 generated by SOD.
In addition, it was discovered that the GDM group's plasma
GSH concentration was lower than that of healthy pregnant
women [31]. This is in line with the results of other research. A
gradual decrease in the activity of glutathione reductase throughout the three trimesters of pregnancy was observed
[32].
Superoxide dismutase1 is an intracellular enzyme that
primarily protects cells against cytosolic generated superoxide.
The expression of cytoplasmic SOD1 is stable and its activity is
often considered as an internal control for SOD gene expression.
SOD1 A251G polymorphisms cause changes in SOD1 enzyme
levels or activities, which can lead to decreased protection
against oxidative stress [33]. This study aimed to demonstrate
that the genotype frequencies of the SOD1 gene (A/G
rs2070424) differed significantly between women with
gestational diabetes and normal pregnant women.
In the Egyptian population, the CC genotype of SOD1
(rs2234694) SNP was more prone to T2DM [34]. It was revealed
that 64.10% of the population has the wild type (AA) SOD1 gene
in the south-south pregnant Nigerian population. 10.26% of the
women had the homozygous genotype and 25.64% of the
population had the heterozygous genotype. In addition, wildtype
SOD1 genes were most prevalent in the population of
women studied [35]. Data regarding the SOD1 gene
polymorphism is available for some populations, including North
Indians, Bangladeshis, the Finns, the Romanians, the New
Zealanders, and the Czechs, but it is lacking among African
populations. There is little information about the frequency or
implications of SOD1 [36].
SOD1 allelic variations (rs17880135) have been linked to the
occurrence of diabetic nephropathy [37]. Another study found
that significant differences between the allele and genotype
frequencies for the SOD1+35A/C polymorphism were
observed in type 1 diabetes mellitus as compared to
controls [38]. In Mexican women, the number of mutant
carriers (GA+GG) of SOD (rs2070424) was substantially
greater in the obese population than in the normal weight group
[39].
However, contradictory results indicated that SOD1
(rs2234694) SNP was not related to cardiovascular disease in
T2DM patients from North Finland. Similarly, it was found that
SOD1+35A/C gene polymorphism may not be associated with
the susceptibility to T2DM among the South Indian population
[40]. Another study has also reported that the (CC) genotype
and C allele were completely absent among the North Indian
population and that there was no association between diabetes
and SOD1+35A/C gene polymorphism [36].
The current study found the SOD1 gene was associated with
blood glucose, MDA, and nitric oxide. The absence of SOD1 is
associated with the development of impaired glucose tolerance
that results from impaired cell dysfunction and reduced cell
volume. ROS reacts with polyunsaturated fatty acids to induce
the release of toxic and reactive aldehyde metabolites such as
MDA, one of the end products of lipid peroxidation. The mutant
SOD1 may have the capacity to catalyze the production of ROS
such as peroxynitrite and also contribute to increased cellular
expression of iNOS synthase.
Conclusion
It was concluded that SOD1-AG (90.0%) genotype were shown
to be the most common in women with GDM. To the best of our
knowledge, this is the first study to show that there is a
significant difference in the frequency of the genotype of SOD1
(A/G rs2070424) in the development of gestational diabetes
mellitus in Egyptian women.
Ethics Approval and Consent to Participate
This study was approved by ethical standards of the
institutional research board, faculty of medicine and Mansoura
university. The patient provided written informed consent.
Competing Interests
The authors declare that they have no competing interests.
Authors’ Contributions
MMM contributed to the methodology, analysis, investigation
and writing original manuscript. FE contributed to the
methodology, acquisition, analysis and investigation. AE
contributed to the methodology, analysis and investigation. AAE
contributed to interpretation of data, supervision, reviewing and
editing of the manuscript. AFA contributed to designing the
work, supervision, reviewing and editing of the manuscript. All
authors have read and approved the manuscript for publication.
Acknowledgment
We express gratitude to all of the obstetrics and gynecology
department, faculty of medicine, Mansoura University for
making it possible to collect maternal blood samples.
References
- Shen Y, Jia Y, Zhou J, Cheng X-Y, Huang H-Y, et al. (2020) Association of gestational diabetes mellitus with adverse pregnancy outcomes: our experience and meta-analysis. Int J Diabetes Dev Ctries 40:357-370 [Crossref] [Googlescholar] [Indexed]
- Park S, Kim MY, Baik SH, Woo JT, Kwon YJ, et al. (2013) Gestational diabetes is associated with high energy and saturated fat intakes and with low plasma visfatin and adiponectin levels independent of prepregnancy BMI. Eur J Clin Nutr 67:196-201 [Crossref] [Googlescholar] [Indexed]
- Murthy KS, Bhandiwada A, Chandan SL, Gowda SL, Sindhusree G (2018) Evaluation of oxidative stress and proinflammatory cytokines in gestational diabetes mellitus and their correlation with pregnancy outcome. Indian J Endocrinol Metab 22:79-84 [Crossref] [Googlescholar] [Indexed]
- Zhu C, Yang H, Geng Q, Ma Q, Long Y, et al. (2015) Association of oxidative stress biomarkers with gestational diabetes mellitus in pregnant women: a case-control study. PLoS One 10:1-12 [Crossref] [Googlescholar] [Indexed]
- Doucette P A, Whitson L J, Cao X, Schirf V, Demeler B, et al. (2004) Dissociation of human copper-zinc superoxide dismutase dimers using chaotrope and reductant: insights into the molecular basis for dimer stability. J Biol Chem 279:54558-54566[Crossref] [Googlescholar] [Indexed]
- Celojevic D, Nilsson S, Behndig A, Tasa G, Juronen E, et al. (2013) Superoxide dismutase gene polymorphisms in patients with age-related cataract. Ophthalmic Genet 34: 140-145 [Crossref] [Googlescholar] [Indexed]
- Aebi H (1984) Catalase in vitro. Meth Enzymol 105:121-126 [Crossref] [Googlescholar] [Indexed]
- Fossati P, Prencipe L, Berti G (1980) Use of 3, 5-dichloro-2-hydroxybenzenesulfonic acid/4-aminophenazone chromogenic system in direct enzymic assay of uric acid in serum and urine. Clin Chem 26:227-231 [Googlescholar] [Indexed]
- Ponti V, Dianzani M, Cheeseman K, Slater T (1978) Studies on the reduction of nitroblue tetrazolium chloride mediated through the action of NADH and phenazine methosulphate. Chem Biol Interact 23:281-291 [Crossref] [Googlescholar] [Indexed]
- Goldberg D, Spooner R (1983) Assay of Glutathione Reductase. Verlag Chemie 258–265
- Satoh K (1978) Method of lipid peroxidation determination in serum. Clin Chim Acta 90:37-43
- Ohkawa H, Ohishi N, Yagi K (1979) Assay for lipid peroxides in animal tissues by thiobarbituric acid reaction. Anal Biochem 95:351-358 [Crossref] [Googlescholar] [Indexed]
- Tsikas D (2005) Review Methods of quantitative analysis of the nitric oxide metabolites nitrite and nitrate in human biological fluids. Free Radic Res 39:797-815 [Crossref] [Googlescholar] [Indexed]
- Weydert CJ, Cullen JJ (2010) Measurement of superoxide dismutase, catalase and glutathione peroxidase in cultured cells and tissue. Nat Protoc 5:51-66[Crossref] [Googlescholar] [Indexed]
- Grupe K, Pfeifer MA, Dannehl F, Liebmann M, Rustenbeck I, et al. (2020) Metabolic changes during pregnancy in glucose-intolerant NZO mice: A polygenic model with prediabetic metabolism. Physiol Rep 8:1-13 [Crossref] [Googlescholar] [Indexed]
- Yuksel S, Yigit AA (2015) Malondialdehyde and nitric oxide levels and catalase, superoxide dismutase, and glutathione peroxidase levels in maternal blood during different trimesters of pregnancy and in the cord blood of newborns. Turk J Med Sci 45:454-459 [Crossref] [Googlescholar] [Indexed]
- Adeniji A O, Oparinde D P (2013) The profiles of lipid peroxidation and anti-oxidant activities in gestational diabetes mellitus and normal pregnancies in Nigerian population. Open J Obstet Gynecol 3:472-476 [Crossref] [Googlescholar] [Indexed]
- Kodliwadmath S, Sadashivadu B, Kodliwadmath M (1989) Serum Malondialdehyde and ceruloplasmin Levels in toxaemia of pregnancy. J Obstet Gynaecol India 5:648-651
- Arribas L, Almansa I, Miranda M, Muriach M, Romero F J, et al. (2016) Serum malondialdehyde concentration and glutathione peroxidase activity in a longitudinal study of gestational diabetes. PLoS One 11:1-13 [Crossref] [Googlescholar] [Indexed]
- Rodrigues F, De Lucca L, Neme WS, Goncalves TL (2018) Influence of gestational diabetes on the activity of delta-aminolevulinate dehydratase and oxidative stress biomarkers. Redox Rep 23:63-67 [Crossref] [Googlescholar] [Indexed]
- Hata T, Hashimoto M, Kanenishi K, Akiyama M, Yanagihara T, et al. (1999) Maternal circulating nitrite levels are decreased in both normal normotensive pregnancies and pregnancies with preeclampsia. Gynecol Obstet Invest 48:93-97 [Crossref] [Googlescholar] [Indexed]
- Brown M A, Tibben E, Zammit VC, Cario GM, Carlton MA (1995) Nitric oxide excretion in normal and hypertensive pregnancies. Pregnancy Hypertens 14:319-326 [Crossref] [Indexed]
- Jr AS, Allman KG, Young D, Redman CW (1997) Elevated levels of serum nitrate, a stable end product of nitric oxide, in women with pre‐eclampsia. Br J Obstet Gynaecol 104: 538-543 [Crossref] [Googlescholar] [Indexed]
- Chaudhary L, Tandon O, Vaney N, Agarwal N (2003) Lipid peroxidation and antioxidant enzymes in gestational diabetics. Indian J Physiol Pharmacol 47:441-446 [Googlescholar] [Indexed]
- Wisdom S J, Wilson R, McKillop J H, Walker JJ (1991) Antioxidant systems in normal pregnancy and in pregnancy-induced hypertension. Am J Obstet Gynecol 165:1701-1704 [Crossref] [Googlescholar] [Indexed]
- Davidge ST, Hubel CA, Brayden RD, Capeless EC, McLaughlin MK (1992) Sera antioxidant activity in uncomplicated and preeclamptic pregnancies. Obstet Gynecol 79: 897-901 [Crossref] [Googlescholar] [Indexed]
- Shang M, Zhao J, Yang L, Lin L (2015) Oxidative stress and antioxidant status in women with gestational diabetes mellitus diagnosed by IADPSG criteria. Diabetes Res Clin Pract 109:404-410 [Crossref] [Googlescholar] [Indexed]
- Ademuyiwa O, Odusoga OL, Adebawo OO, Ugbaja R (2007) Endogenous antioxidant defences in plasma and erythrocytes of pregnant women during different trimesters of pregnancy. Acta Obstet Gynecol Scand 86:1175-1182 [Crossref] [Googlescholar] [Indexed]
- Djordjevic A, Spasic S, Jovanovic-Galovic A, Djordjevic R, Grubor-Lajsic G (2004) Oxidative stress in diabetic pregnancy: SOD, CAT and GSH-Px activity and lipid peroxidation products. J Matern Fetal Neonatal Med 16:367-372 [Crossref] [Googlescholar] [Indexed]
- Goth L, Toth ZN, Tarnai I, Berces M, Torok P, et al. (2005) Blood catalase activity in gestational diabetes is decreased but not associated with pregnancy complications. Clin Chem 51:2401-2404 [Crossref] [Googlescholar] [Indexed]
- Jamil D, Al-Aubaidy H, Smith L, Jelinek H (2014) Oxidative DNA damage in gestational diabetes mellitus: correlation with antioxidants in an Iraqi cohort. Int J Biochem Res Rev 4:410-419 [Crossref] [Googlescholar] [Indexed]
- Patil S B, Kodliwadmath M V, Kodliwadmath SM (2007) Study of oxidative stress and enzymatic antioxidants in normal pregnancy. Indian J Clin Biochem 22:135-137 [Crossref] [Googlescholar] [Indexed]
- Silig Y, Tas A, Sahin-Bolukbasi S, Caglayan G,Sari I (2017) Superoxide Dismutase 1 (SOD 1) A251G Polymorphism. Turk J Biochem 42:181-185 [Crossref] [Googlescholar] [Indexed]
- Ghattas MH, Abo-Elmatty DM (2012) Association of polymorphic markers of the catalase and superoxide dismutase genes with type 2 diabetes mellitus. DNA Cell Biol 31:1598-1603 [Crossref] [Googlescholar] [Indexed]
- Imaobong A, Ubong A, Glory S, Offiong E, Ndodo N (2017) Genotyping of A251g, copper-zinc superoxide dismutase (Cuznsod) polymorphism, in term pregnant women In Uyo, South-South, Nigeria. Int J biotechnol biochem 3:19-22 [Googlescholar]
- Vats P, Sagar N, Singh T P, Banerjee M (2015) Association of Superoxide dismutases (SOD1 and SOD2) and Glutathione peroxidase 1 (GPx1) gene polymorphisms with type 2 diabetes mellitus. Free Radic Res 49:17-24 [Crossref] [Googlescholar] [Indexed]
- Mohammedi K, Maimaitiming S, Emery N, Bellili-Munoz N, Roussel R, et al. (2011) Allelic variations in superoxide dismutase-1 (SOD1) gene are associated with increased risk of diabetic nephropathy in type 1 diabetic subjects. Mol Genet Metab 104:654-660 [Crossref] [Googlescholar] [Indexed]
- Flekac M, Skrha J, Hilgertova J, Lacinova Z, Jarolimkova M (2008) Gene polymorphisms of superoxide dismutases and catalase in diabetes mellitus. BMC Med Genet 9:30-39 [Crossref] [Googlescholar] [Indexed]
- Hernandez-Guerrero C, Hernandez-Chavez P, Romo-Palafox I, Blanco-Melo G, Parra-Carriedo A, et al. (2016) Genetic polymorphisms in SOD (rs2070424, rs7880) and CAT (rs7943316, rs1001179) enzymes are associated with increased body fat percentage and visceral fat in an obese population from Central Mexico. Arch Med Res 47:331-339 [Crossref] [Googlescholar] [Indexed]
- Nithya K, Angeline T, Isabel W, Asirvatham A (2016) SOD1 gene+35A/C (exon3/intron3) polymorphism in type 2 diabetes mellitus among south indian population. Genet Res Int 2016:1-5 [Crossref] [Googlescholar] [Indexed]
Citation: Madkour S, Abd El-Aziz AAF, El-Senduny FF, Elsaid AM, El-Refaey AAA (2023) Gene Polymorphism of Superoxide Dismutase in Gestational Diabetes Mellitus in Pregnant Women. Ann Clin Lab Res Vol:11 No:01