Key words
Aeromonas, RAPD, Food animals
Introduction
Aeromonas spp. has become increasingly recognized as enteric pathogens. These organisms cause acute diarrhoea in children [1] and adults [6] and sporadic diarrhoea or dysentery in those older than 60 years, which can be severe and even life threatening [4-5]. However, today, these are also responsible for causing gastroenteritis outbreaks in humans and traveler’s diarrhea [19].
The spectrum of infectious diseases caused by Aeromonas species includes gastrointestinal infections as well as extra intestinal infections such as cellulites, wound infections, septicemia, urinary tract infection and hepatobiliary and ear infections [15]. Virulence of Aeromonas spp. is multifactorial and incompletely understood. Factors contributing to virulence include toxins, proteases, hemolysins, lipases, adhesins, agglutinins, and various hydrolytic enzymes [7,2]. These virulence factors are useful in distinguishing between potentially pathogenic and non-pathogenic strains. Some investigators observed that Aeromonas induced gastroenteritis is due to an enterotoxin which is cytotoxic in nature [16,14] while others reported aerolysin to be the main virulence factor involved in intestinal disorders. About 6.5% of diarrhoeal cases in the southern part of India have been attributed to Aeromonas [10], which indicates an urgent need for information on the casual role of this pathogen in other parts of the country.
Aeromonas associated gastroenteritis is probably under diagnosed due to the lack of recognition of its significance, confusion over its taxonomy and the difficulty for a laboratory to routinely identify isolates with virulence-associated properties, such as enterotoxin production and entero-invassiveness [8]. According to the International Commission on Microbiological specifications for Food 1996, many classical procedures for the detection of Aeromonas spp. were found to be laborious and time consuming or not allowing quantitative assessment of these organisms, thus indicating the need for a reliable, universal and standard method.
Materials and Methods
Bacterial strains and culture conditions
A total of 332 samples of which 38 isolates were identified by the 16S rRNA technique were included in the study. The isolates were grown on ADA (Ampicillin Dextrin Agar (HI-MEDIA Laboratories, Mumbai, India) at 37°C for 18-24 hours. Of the 38 isolates, 35 (92.10%), 2 (5.26%) and 1 (2.63%) were recognized as A. hydrophila, A. sobria and A. caviae. All the strains were dominantly environmental isolates in our collection. The details of the samples collected have been summarized in the following Table one.
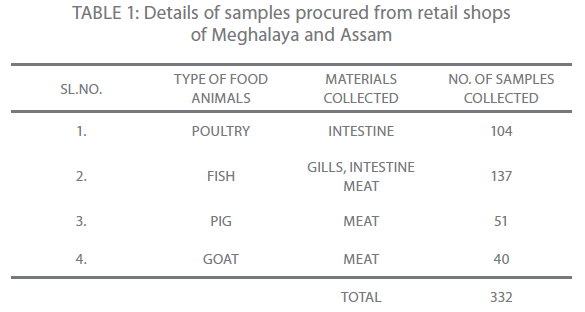
Table 1: Details of samples procured from retail shops of Meghalaya and Assam
Biochemical studies
The following biochemical tests were done in all strains according to conventional protocols: esculin hydrolysis, citrate utilization, motility, indole production, and acid production from rhamnose, sorbitol, lactose, D-sucrose, and salicin. Test samples were incubated under the same conditions as used for bacterial growth. All tests were carried out in duplicate, and appropriate positive and negative controls were included.
Molecular typing of isolates by PCR
The use of PCR assay for molecular typing revealed that all the 38 isolates belonged to Aeromonas spp. and were positive for 16S rRNA (100%), ahh1 (60.52%), asa1 (42.10%), A. hydrophila aerA (13.15%) and AHCYTOEA/aerA (5.26%). In the present investigation it was noted that aerolysin producing toxin genes were the most prevalent irrespective of their geographical locations and the most common single gene carried among all the isolates examined was ahh 1 (60.52%). The results recorded in this study are in agreement with those reported by Wang et al. 2003 [18] in the multiplex PCR.
Isolation of genomic DNA
Colonies of Aeromonas spp. grown on Ampicilin Dextrin agar at 37°C for 24 hr. were scraped off and suspended in 0.85% NaCl. After centrifugation, the pellet was resuspended in a lysis buffer and incubated after adding protease K. The DNA was extracted using the Promega (USA), DNA isolation Kit according to the procedure established by the manufacturer. The quality and quantity of the DNA were determined spectrophotometrically at 260 nm. The extracted DNA was stored at 2-8oC for further use.
Random amplification of polymorphic DNA
Primers for RAPD [12] used in the study are presented in Table 2. Amplification was performed using a thermocycler (iCycler, BIORAD, USA). The 50 μl reaction contained 10 x NH4 buffer, 4.0 mM Mgcl2, 200 μM of dNTPs, 2U Taq DNA polymerase and 20 ng of genomic DNA. Amplification conditions were 35 cycles of 10 sec. at 94°C, 30 sec. at 37°C, 60 sec. at 72°C and a final extension of 5 min. at 72°C. The amplification products were analyzed in 1% agarose (Promega, USA) gel. The gel was prepared by dissolving agarose in 1X Tris – acetate (TAE) buffer (Genei, Bangalore). Same buffer was used for electrophoretic run. A total of 5μl of each amplicons and 1μl marker DNA (100bp DNA ladder mix; MBI Fermentas, USA) were mixed separately with 1μl of 6X gel loading dye (MBI Fermentas, USA) and loaded in the wells of the gel. Electrophoresis was carried out in Mini plus horizontal (GENE Mate® gel system, UK) electrophoretic apparatus at a constant voltage of 60 V for 1 hour and 20 min. or until the second dye marker had run 3/4th of the gel. Then the gel was stained in ethidium bromide (Pharmacia Biotech, Sweden) @ 0.4 μg/ml in distilled water solution for 10-15 min. and was visualized in gel documentation system (Gel Logic 100 Imaging System, Biostep) and photographed.
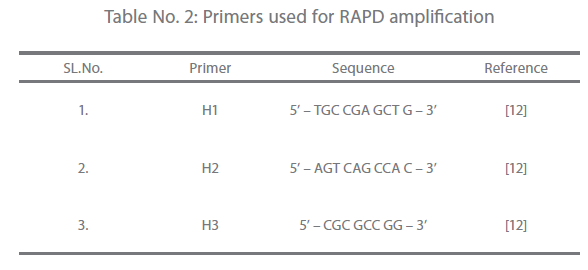
Table 2: Primers used for RAPD amplification
Results
Biochemical studies
Biochemical characterization of the isolates revealed that heterogenecity existed between the isolates. All the 38 isolates screened for primary characterization were tested positive for oxidase and catalase test. The Gram stained Aeromonas isolates showed Gram-negative bacilli appearing as short rods. Further confirmation test which characterized the Aeromonas isolates upto species level were performed using standard protocols. Some variability was observed with regard to VP test and gelatin liquefaction. However, biochemical reactions were found to deviate from the ideal phenotype of each isolate. Of the 38 isolates, 35 (92.10%), 2 (5.26%) and 1 (2.63%) were recognized as A. hydrophila, A. sobria and A. caviae.
RAPD analysis
In the present study, out of 20 random primers of OPH series, OPH-1, OPH-2 and OPH-3 were found to produce the most reproducible and scorable amplicon profiles. The amplicon sizes ranged from 250 bp to 2.9 Kb with a common >250 bp fragments in all the isolates (Fig. 1 and 2). The dendogram from the overlaid graphs arising from the RAPD profiles of Aeromonas isolates with primer OPH-1, OPH-2 and OPH-3 revealed that similarity ranged upto 75% in all the samples. Four clusters were observed comprising of C18 and FG16; FI9 and C3; FM4 and PM2. Sample PM2 exhibited 12% similarity, C18, FG16, FI9 and C3 exhibited 28% similarity where as sample FM4 exhibited 44% similarity. Also, PM2 was found to be completely different from other clusters (Fig 3).
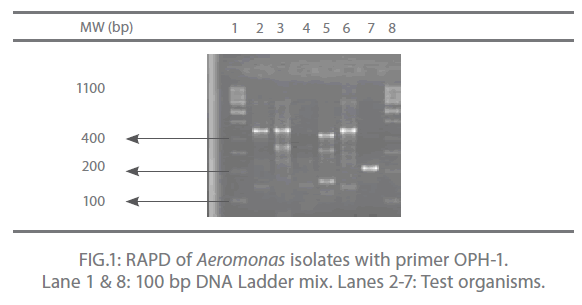
Figure 1: RAPD of Aeromonas isolates with primer OPH-1. Lane 1 & 8: 100 bp DNA Ladder mix. Lanes 2-7: Test organisms.
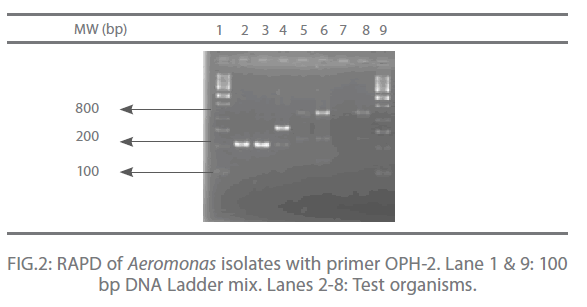
Figure 2: RAPD of Aeromonas isolates with primer OPH-2. Lane 1 & 9: 100 bp DNA Ladder mix. Lanes 2-8: Test organisms.
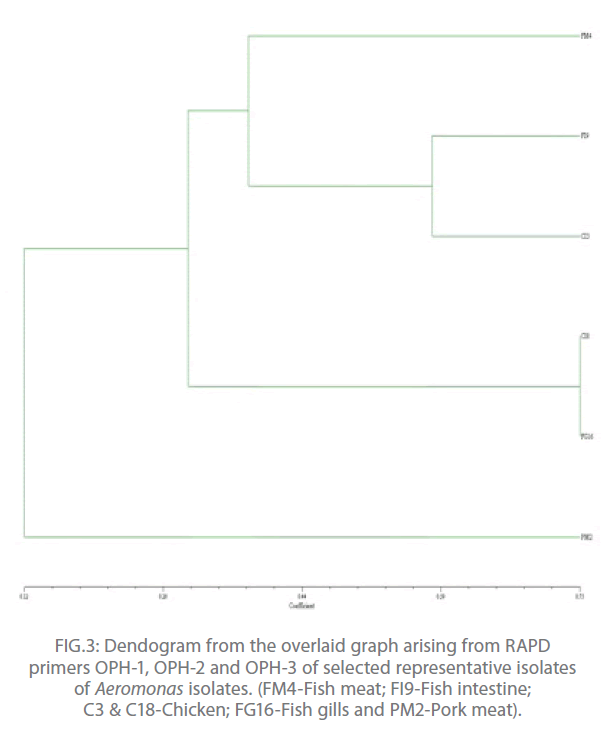
Figure 3: Dendogram from the overlaid graph arising from RAPD primers OPH-1, OPH-2 and OPH-3 of selected representative isolates of Aeromonas isolates. (FM4-Fish meat; FI9-Fish intestine; C3 & C18-Chicken; FG16-Fish gills and PM2-Pork meat).
Discussion
Characteristic biochemical reactions revealed that all the A. hydrophila isolates hydrolyzed aesculin thereby producing brownish black colour of the medium and the A. sobria isolates exhibited negative results by not hydrolyzing aesculin. Furthermore, production of acid from Arabinose was observed both in A. hydrophila and A. sobria isolates but the A. caviae isolate exhibited negative acid production. Some investigators [3] identified Aeromonas upto the genomospecies level by the use of citrate and production of acid from which it enabled them to separate the members of A. hydrophila complex and further reported that these pathogenic genomospecies should be regarded as an important threat to public health. Since Aeromonas genus is a heterogeneous group, some differences have been observed.
Since the identification of Aeromonas spp. often requires the use of non-conventional biochemical assays that are time consuming and often require long incubation period before the final results can be recorded, attempts have been made to find out the minimal identifying characteristics for the use in clinical laboratories. Based on observations recorded in the present investigation and that of reported positivity of different characteristics [9,2], the use of three tests for the identification of Aeromonas spp. with about 90% accuracy has been recommended. These tests include aesculin hydrolysis, Voges- Proskauer and gas from glucose. Strains which are aesculin negative, VP positive and gas from glucose positive can be identified as A. hydrophila, where as A. caviae are negative for gas from glucose and aesculin hydrolysis and do not ferment glucose but positive for VP. In contrast A. sobria ferments glucose and hydrolysis aesculin and maybe VP negative or positive. But in contrary to this, Martínez-Murcia et al., 2005 [11] reported that none of the biochemical tests evaluated in their present study were able to separate two species.
The RAPD assay is an effective way of distinguishing isolates of various pathogenic anaerobic bacteria for epidemiological investigation and also for tying of isolates from different disease outbreaks [17]. In the present study, out of 20 random primers of OPH series, OPH-1, OPH-2 and OPH-3 were found to produce the most reproducible and scorable amplicon profiles. The amplicon sizes ranged from 250 bp to 2.9 Kb with a common >250 bp fragments in all the isolates (Fig. 1 and 2). Similar results were also observed by O’hIci et al. 2000 [12]. They reported close genetic matrix similarity among the tropical strains using the OPH series primer (H1, H2 and H3). All the representative isolates in the present investigation revealed similarity upto 75% (Fig 3). This was in conformity with the similarity reported by other investigators [12-13]. Such differences may well be related to the source, frequency and type of Aeromonas isolates encountered in different geographical areas. The present study has established that when an appropriately chosen set of primers is employed, the RAPD analysis provides an alternative, rapid, reproducible and powerful genomic typing for the Aeromonas species.
In conclusion, the RAPD revealed that amplicon sizes ranged from 250 bp to 2.9 Kb with a common >250 bp fragments in all the isolates and 75% similarity was observed in all the representative isolates. Presence of four clusters were observed comprising of C18 and FG16; FI9 and C3; FM4 and PM2.
Acknowledgment
The first author is thankful to the university authorities for providing financial assistance in the form of scholarship.
278
References
- Agger WA (1986) Diarrhoea associated with Aeromonashydrophila. Pediatr Infect Dis 5(Suppl.): 105-108.
- Abbott LS, Cheung WKW, Bystrom-Kroske S, Malekzadeh T, JandaMJ (1992) Identification of Aeromonasstrains to the genospecies level in the clinical laboratory. J ClinMicrobiol 30(5): 1262-1266.
- Borrell N, FiguerasJM, Guarro J (1998) Phenotypic identification of Aeromonasgenomospecies from clinical and environmental sources. Can J Microbiol 44(2):103-108.
- Champsaur H, Andermont A, Matheiu D, Rottman E, Auzepy P (1982) Cholera-like illness due to Aeromonassobria. J Infec Dis 145: 361-366.
- Echeverria P, Blacklow NR, Sanford LB, Cukor GC (1981) Travellers’diarrhoea among American peace corps volunteers in rural Thailand. J Infec Dis 143: 767-771.
- Gracey M, Burke V, Robinson J (1982) Short reports- Aeromonasspp. in traveller’sdiarrhoea. British Med J 289:658.
- JandaJM, Abott SL (1996) Human pathogens. In: Austin B et al., eds. The genus Aeromonas.London, Wiley. pp. 151-173.
- JandaJM, GuthertzLS, Kokka RP, Shimada T (1994) Aeromonasspecies in septicemia: laboratory characteristics and clinical observations. Clinical Infectious Diseases 19: 77–83.
- Joseph SW, Colwell RR, McDonell MT (1987) Taxonomy, ecology, isolation and identification: Aeromonastaxonomy. Experrientia 43: 349-550.
- Komathi AG, Ananthan S, AlavandiSV (1998) Incidence and enteropathogenecity of Aeromonmas species associated with childhood gastroenteritis in Chennai (Madras), India. Jpn J Med SciBiol 51: 1-12.
- Murcia-Martinez JA, Soler L, SaavedraJM, Chacon RM, Guarro J, et al. (2005) Phenotypic, genotypic, and phylogenetic discrepancies to differentiate Aeromonassalmonicida from A. bestiarum. IntMicrobiol 8: 259-269.
- O’hIci B, Oliver G, Powell R (2000) Genetic diversity of the fish pathogen Aeromonassalmonicidademonstrated by random amplified polymorphic DNA and pulsed – field gel electrophoresis analysis. Dis Aquat Org 39: 109-119.
- Redondo NP, Jarero RJ, Figueroa ALJ (2004) Antibiotic resistance and presence of plasmids in: Aeromonashydrophila, Vibrio fluvialis and Vibrofurnissi isolated from Carassiusauratusauratus. Vet Mex 35(1): 1-10.
- StelmaGN, Johnson CH, Spaulding P (1986) Evidence for the direct involvement of ß-hemolysin in Aeromonashydrophilaenteropathogenecity. CurrMicrobiol 14: 71-77.
- Vila J, Ruiz J, Gallardo F, Vargas M, Soler L, et al. (2003) Aeromonasspp. and traveller’sdiarrhoea: Clinical features and antimicrobial resistance. EmergInfec Dis 9(5): 552-555.
- Wadstrom T, Ljungh A, Wretling B (1976) Enterotoxin, haemolysin and cytotoxic protein in Aeromonashydrophilafrom human infections. ActaPatholMicrobiolScand 8: 112-114.
- Wang G, Tyler DK, Munro KC, Johnson MW (1996) Characterization of cytotoxic, haemolyticAeromonascaviaeclinical isolates and their identification by determining presence of a unique haemolysin. J ClinMicrobiol 34(12): 3203-3205.
- Wang G, Clark GC, Liu C, Pucknell C, Munro KC, et al. (2003) Detection and characterization of the haemolysin genes in Aeromonashydrophilaand A. sobria by multiplex PCR. J ClinMicrobiol 41(3): 1048-1054.
- Yamada S, Matsushita S, Dejsirilert S, Kudoh Y (1997) Incidence and clinical symptoms of Aeromonasassociated traveller’s diarrhea in Tokyo. Epidemiol Infect 119:121-126.










