Introduction
The majority of acute dentoalveolar abscesses involve dental pulp necrosis [1,2]. Although vital dental pulp is normally sterile, deep caries and serious dental trauma often result in pulp infection with bacteria residing within the mouth. Infected regions of pulp will necrotize, and subsequently, bacteria may diffuse from the necrotic pulp into the periapical tissues including the alveolar bone. Although the tissues are colonized with a number of bacteria, the host defense systems usually maintain homeostasis between host and bacteria. However, once microbial homeostasis is disrupted, bacterial numbers increase and invade the tissues, accompanied by acute inflammation. Common clinical signs and symptoms include tooth pain and gingival swelling. Extensive facial and cervical swelling may occur in advanced cases, often accompanied by malaise and fever. Although rare, infection can cause serious complications including airway obstruction and sepsis [1,2].
Acute dentoalveolar abscess is usually polymicrobial, primarily involving strict anaerobes and oral streptococci [1-7]. Experimental studies have demonstrated that an enhancement of bacterial virulence occurs in mixed infection of these organisms [8-12]. Consequently microbial synergy is considered a possible etiologic factor of infection [4].
The genus Prevotella, is comprised of species that are strict anaerobic Gram-negative bacilli. Members of this genus are a major component of the human microflora at various body sites including the mouth [13], and they are often predominant in acute dentoalveolar abscess [1-7]. The Prevotella genus consists of more than 10 species, which are roughly divided into black-pigmented and non-pigmented types by the color of colonies forming on blood agar [13]. Pigmented Prevotella species frequently involved in dentoalveolar abscess include P. intermedia, P. nigrescens, P. melaninogenica and P. loescheii [7,14]. Prevotella oralis, P. oris and P. buccae are the non-pigmented Prevotella species that are most frequently isolated from the infection [7,14]. Both pigmented and non-pigmented Prevotella species have the potential to produce purulent infection in an experimental animal model [15]. Thus Prevotella are known to play a significant role in development of acute dentoalveolar abscess.
It has widely been accepted that strains of each bacterial species involved in the infection of an individual are genetically homogeneous [16]. However, scientific evidence to support this hypothesis is only available for a limited number of infections. In infections within the mouth, a high degree of genetic similarity between Porphyromonas gingivalis strains recovered from periodontitis of same individual has been reported [17]. However, there has been no investigation to determine the genetic diversity of strains within dentoalveolar abscess of individual patients. Furthermore, with regards to other human infections involving Prevotella, no study has been conducted to address this hypothesis.
As dentoalveolar abscess occurs opportunistically, it can reasonably be hypothesized that the bacteria of the same genotype acts both as commensals and causative agents of infection. However, little is known concerning the genetic homogeneity of these opportunistic pathogens during commensal existence and infection.
The purpose of this study was therefore to investigate the genetic diversity of Prevotella strains recovered from dentoalveolar abscess. The study also determined the genetic relatedness between clinical and commensal oral isolates from the same patients.
Methods
Specimens. Pus and plaque specimens were obtained from a total of 31 patients with acute dentoalveolar abscess (18 males and 13 females, mean age 54.5 years) attending oral and maxillofacial surgery clinics at Kanazawa University Hospital (Kanazawa, Japan) and Noto General Hospital (Nanao, Japan) between March 2007 and December 2009. All patients had tenderness and swelling of the vestibule site around the involved tooth, and mild to moderate facial swelling. All patients were however free of any serious complications including sepsis and airway obstruction. No patient had a history of being immuno-compromised or having received medication that could significantly influence immunity. Pus was obtained from the obstructive abscess by aspiration. Dental plaque was obtained from around the gingival margin of three noninfected teeth that exhibited healthy periodontal tissue. These teeth were chosen randomly in patients and collected using a fine dental instrument immediately after obtaining the pus specimen. Informedconsent for this study was obtained from all patients.
Culture and identification of isolates. Pus and plaque samples were inoculated on to Brucella HK agar (Kyokuto, Tokyo, Japan) containing 5% (v/v) sheep blood for up to 72 h at 37°C under aerobic and anaerobic conditions. Each specimen was also incubated on blood agar containing paromomycin (75mg/L, Pfizer, Tokyo, Japan) and vancomycin (2.5mg/L, Shionogi, Osaka, Japan) for up to 72 h at 37°C under anaerobic conditions to selectively isolate strictly anaerobic Gram-negative bacilli. Six to 10 presumptive Prevotella colonies per specimen were selected from each primary culture plate and subcultured individually on fresh agar plates. Isolates from pus and plaque were identified by conventional methods [5,13]. Identification of Prevotella strains was achieved at species level using Rap ID ANA II system (Remel, Lenexa, KS)[14].
Typing. In cases where the same Prevotella species was recovered from both pus and plaque of individual patients, the strains were genotyped using a random amplified polymorphic DNA (RAPD) assay [18] and the antimicrobial susceptibility of these isolates was also determined. With regards to the RAPD assay, colonies of each subcultured strain were resuspended in saline. Bacterial DNA was extracted and purified using the DNeasy Blood & Tissue Kit (Qiagen, Tokyo, Japan). The RAPD assay was performed using Ready-To-Go RAPD Analysis Beads (GE Healthcare Japan, Tokyo, Japan). Primer 2 (5’-GGTGCGGGAA-3’) and primer 3 (5’-CCCGTCAGCA-3’) for amplification were selected by comparing the amplification patterns of each of six primers offered by the manufacturer. The PCR was undertaken in accordance with manufacturer’s recommendations. Analysis of each PCR product was performed by gel electrophoresis with 1.5% agarose gel (Nacalai tesque, Kyoto, Japan) at 100 V/cm for 1 h and subsequent ethidium bromide staining. The banding patterns of tested strains appearing on the gel image were visually compared. All experiments were repeated in triplicate and on three separate occasions to confirm reproducibility.
For isolated strains, minimum inhibitory concentrations (MICs) for amoxicillin (Astellas, Tokyo), amoxicillin/clavulanate (2:1, GlaxoSmith- Kline, Middlesex, UK), cefaclor (Shionogi), ceftazidime (GlaxoSmith- Kline), erythromycin (Shionogi), azithromycin (Pfizer), minocycline (Takeda, Tokyo), clindamycin (Pfizer), levofloxacin (Daiichi-Sankyo, Tokyo), and metronidazole (Shionogi) were determined using the agar dilution method recommended by the Clinical and Laboratory Standards Institute [19]. Bacteroides fragilis ATCC 25285 and Bacteroides thetaiotaomicron ATCC 29741 were used as quality control strains in each test.
Results
The results of microbiological examination of pus and plaque are presented in Table 1. Prevotella species were isolated from pus specimens in 18 (58%) of 31 patients and recovered from the plaque in 20 (65%) of 31 patients. The pus isolates included P. buccae (10 strains), P. loescheii (4 strains), P. melaninogenica (3 strains), P. intermedia/P. nigrescens (3 strains), P. oralis (2 strains), P. denticola (1 strain), P. disiens (1 strain), and unidentified Prevotella species (4 strains). With regards to plaque, the isolates included P. buccae ?14 strains), P. melaninogenica (6 strains), P. intermedia/P. nigrescens (5 strains), P. loescheii (3 strains), P. oralis (3 strains), P. denticola (3 strains), P. disiens (1 strain), P. oris (1 strain), and unidentified Prevotella species (11 strains).
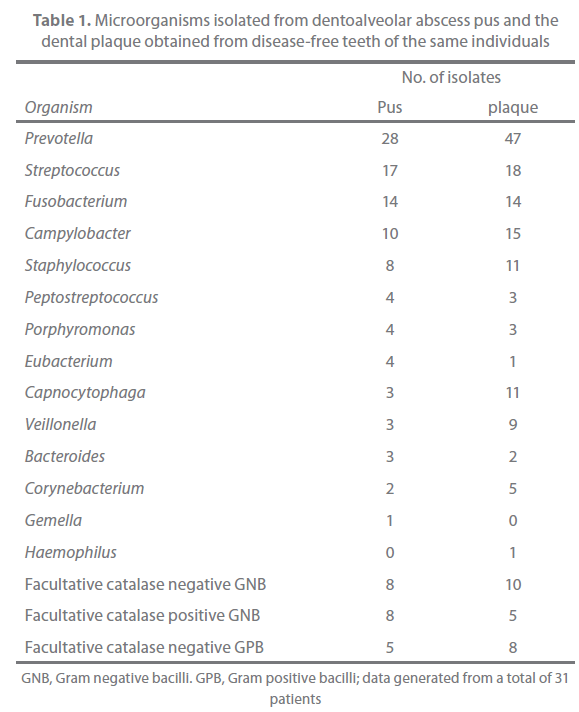
Table 1. Microorganisms isolated from dentoalveolar abscess pus and the dental plaque obtained from disease-free teeth of the same individuals
Of the 31 patients, 9 yielded the same species of Prevotella both in pus and plaque (Table 2). These strains were typed with the RAPD assay and examined for their antimicrobial susceptibility. All strains with same banding pattern exhibited the same or nearly the same MICs for the tested antibiotics (Figure 1). All strains exhibiting different banding patterns were confirmed to have different antimicrobial susceptibility. The results are summarized in Table 3. Multiple genotypes were evident from pus in all of the 9 patients and for the plaque of 3 patients. The pus and plaque specimens from each of 6 patients (patients A, B, C, E, H, and I) had species of the same genotype. Interestingly, although all genotypes of pus isolates were also seen in plaque isolates in 2 of the 6 patients (patients A and C), pus from the remaining 4 patients (patients B, E, H, I) harbored more than one genotype that was not detected in plaque. No genetic agreement between pus and plaque strains occurred in 3 patients (patients D, F, and G).
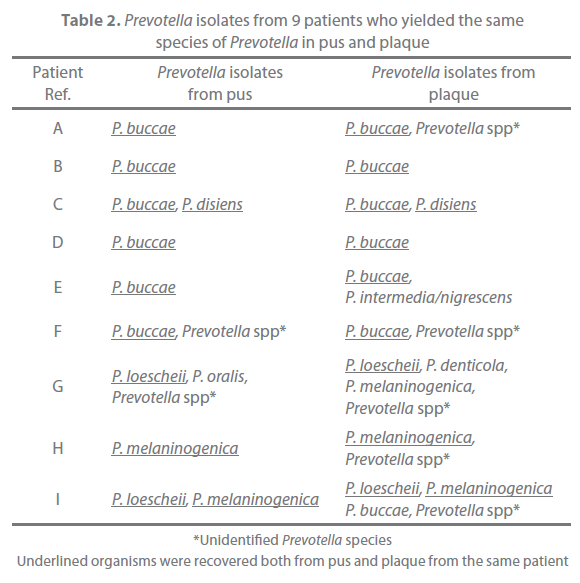
Table 2. Prevotella isolates from 9 patients who yielded the same species of Prevotella in pus and plaque
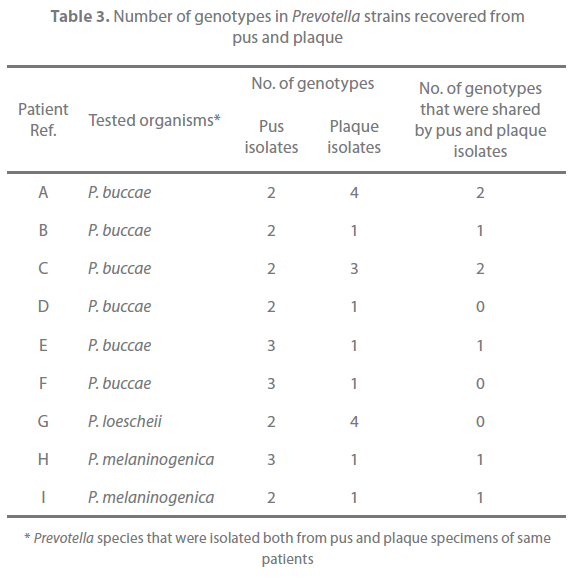
Table 3. Number of genotypes in Prevotella strains recovered from pus and plaque
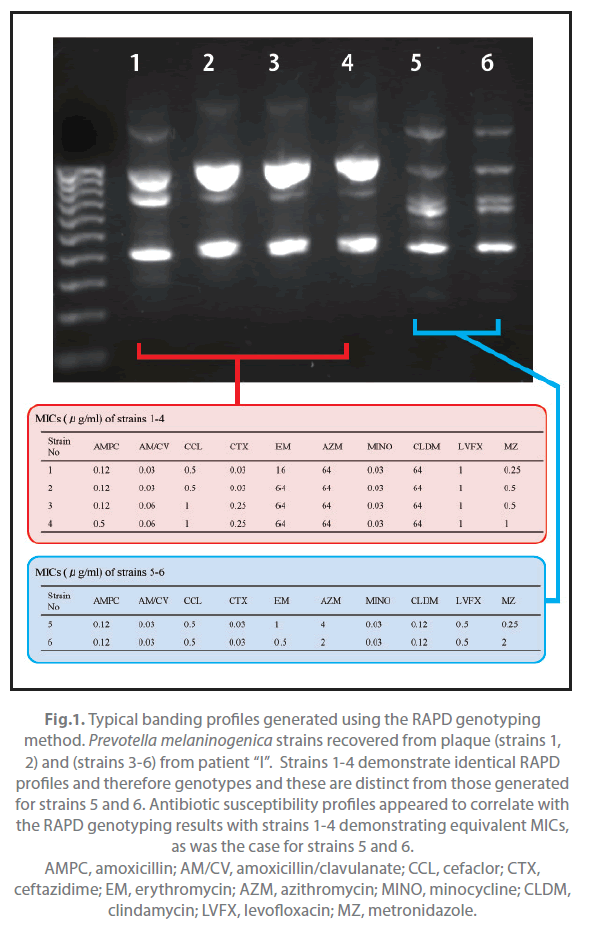
Figure 1. Typical banding profiles generated using the RAPD genotyping method. Prevotella melaninogenica strains recovered from plaque (strains 1, 2) and (strains 3-6) from patient “I”. Strains 1-4 demonstrate identical RAPD profiles and therefore genotypes and these are distinct from those generated for strains 5 and 6. Antibiotic susceptibility profiles appeared to correlate with the RAPD genotyping results with strains 1-4 demonstrating equivalent MICs, as was the case for strains 5 and 6. AMPC, amoxicillin; AM/CV, amoxicillin/clavulanate; CCL, cefaclor; CTX, ceftazidime; EM, erythromycin; AZM, azithromycin; MINO, minocycline; CLDM, clindamycin; LVFX, levofloxacin; MZ, metronidazole.
Discussion
In recent studies using appropriate methods of specimen collection and culture, Prevotella was found to be the most common isolate from acute dental abscess [5,7,20]. High prevalence of bacteria of this genus has also been noted by investigations using molecular techniques [21,22]. In contrast, the incidence of individual Prevotella species varies in published studies, despite similar species being recovered. This discrepancy might be associated with geographical location of study, nature of subjects, and the employed study method [13,23,24].
Prevotella species are readily susceptible to clindamycin and metronidazole [14], whilst 30-80% of clinical isolates are reported to be resistant to penicillin [5,25,26]. Moreover, prevalence of penicillin-resistant Prevotella isolates from dental abscess appears to be increasing as indicated in previous studies by our group [27]. In the present study, although all of the strains obtained from the 9 patients were susceptible to amoxicillin (data not presented), a greater number of clinical isolates is required to address actual status of susceptibility in Prevotella strains involved in dental infection.
The management of acute dentoalveolar abscess consists primarily of surgery, including drainage through incision and tooth extraction, antimicrobial therapy, and medical supportive care [1,2]. Administered antibiotic is usually chosen empirically [1,2], and members of the penicillin class are often recommended as the first-line agent [1,2]. However, antibiotic selection based on data from microbiological examination is highly recommended in circumstances where the expected outcome of therapy has not been obtained, as well as in cases of severe infection, and in immuno-compromised patients [1,2]. The hypothesis that a single genotype of a species is involved in any given bacterial infection is based on interpretation of laboratory data, with very few types of infection having been demonstrated to involve multiple genotypes. One notable exception is the recovery of multiple genotypes of Streptococcus pyogenes from pyoderma skin sores of the same patients [28].
Unexpectedly, however, the typing results of clinical strains in this study indicate that multiple subtypes of species are present at dentoalveolar abscess infection sites with (Table 3). Multiple subtypes also appeared to exist for commensal Prevotella strains. Fukui et al [29] reported that multiple subtypes of P. nigrescens were evident in 57% of the tested 14 dental plaques indicating that a wide diversity of genotypes of commensal Prevotella strains may exist within the mouth of an individual. In the case of Fusobacterium nucleatum, another opportunistic pathogen for dentoalveolar abscess, more than one genotype has been recovered from the healthy mouth of an individual [30]. Since the dentoalveolar abscess is an opportunistic infection, it is perhaps not surprising that heterogeneous Prevotella strains with different genetic and antimicrobial susceptibility characteristics are involved. This in itself would be a matter of concern for clinicians who could receive microbiological test results for only one strain, thereby not reflecting any distinct antibiotic susceptibility variation that could be present at the infection site. Although further study is required, the results of this study have significance in our current understanding of the bacteriology of dental purulent infections.
As dental plaque is a biofilm formed by bacteria colonizing dental surfaces, this material was considered appropriate to obtain commensal strains. In this study, plaque was collected from teeth associated with a healthy periodontal tissue, and these sites were selected since bacteria in subgingival plaque of teeth with periodontitis are associated with the infection itself [31]. In this study, pus and plaque shared the same strain genotype in 6 of 9 cases (Table 3). In 2 of the 6 patients, all strains genotypes in pus isolates were seen in plaque isolates. Since dentoalveolar abscess is an opportunistic infection, it would be expected that the same strains act as commensals and causative agents of infection within an individual. Interestingly, for the other 4 patients, whilst a shared Prevotella genotype was evident for pus and plaque specimens, genotypes restricted to the pus specimens were evident. Moreover, no genetic agreement between pus and plaque strains occurred in 3 cases. This might indeed be a reflection of a wide genetic heterogeneity for both clinical and commensal Prevotella strain populations. However, the discrepancy between the recovered genotypes from commensal and infected sites might also relate to the different selection pressures at the respective locations. Interestingly, it has been demonstrated that Prevotella isolates from dental abscess are more proteolytic compared with those from dental plaque obtained from healthy volunteers [32]. In the case of P. gingivalis, strains carrying specific genes involved in virulence have a high frequency of recovery from severe forms of periodontitis [33]. It may therefore follow that only selected strains with greater virulence suitably adapt to the altered environment of an abscess and thus become causative agents of infection, although further study is necessary to investigate this.
Acknowledgements
We wish to thank Miss Hayashi E (Asanogawa General Hospital), Dr Kato K (Kanazawa University Hospital), Dr. Yoshizawa K (Kanazawa University Hospital), Dr. Kitahara H (Kanazawa University Hospital), Dr. Terai K (Terai Dental Practice), and Dr. Hase T (Noto General Hospital) for their kind cooperation.
Funding
This study was supported with grains-in-aid for scientific research (No. 19791502), the Ministry of Education, Science, Culture, Sports and Science and Technology, Japan.
Competing interests
No competing interests to be declared.
272
References
- Kuriyama T, Lewis MAO, Williams DW (2010) Infection of oral and maxillofacial regions. In: Andersson L, KahnbergKE, Pogrel MA, editors. Oral and maxillofacial surgery. Oxford: Wiley-Blackwell. pp. 467-582.
- Peterson LJ (1998) Principles of management and prevention of odontogenic infections. In: Peterson LJ, Ellis E, Hupp JR, Tucker MR, editors. Contemporary oral and maxillofacial surgery. St. Louis: Mosby. pp. 392-417.
- Robertson D, Smith AJ (2009) The microbiology of the acute dental abscess. J Med Microbiol 58:155-162.
- Dahlén G (2002) Microbiology and treatment of dental abscesses and periodontal-endodontic lesions. Periodontol 2000 28: 206-239.
- Kuriyama T, Karasawa K, Nakagawa K, Saiki Y, Yamamoto E, et al (2000) Bacteriologic features and antimicrobial susceptibility in isolates from orofacialodontogenic infections. Oral Surg Oral Med Oral Pathol Oral RadiolEndod 90: 600-608.
- Lewis MAO, MacFarlane TW, McGowan DA (1990) A microbiological and clinical review of the acute dentoalveolar abscess. Br J Oral MaxillofacSurg 28: 359-366.
- Brook I, Frazier EH, Gher ME (1991) Aerobic and anaerobic microbiology of periapical abscess. Oral MicrobiolImmunol 6:123-125.
- Van Dalen PJ, van Deutekom-Mulder EC, de Graaff J, van SteenbergenTJM (1998) Pathogenicity of Peptostreptococcus micros morphotypes and Prevotellaspecies in pure and mixed culture. J Med Microbiol 47: 135-140.
- Kuriyama T, Nakagawa K, Kawashiri S, Yamamoto E, Nakamura S, et al (2000) The virulence of mixed infection with Streptococcus constellatusand Fusobacteriumnucleatumin a murine orofacial infection model. Microbes Infect 2: 1425-1430.
- Brook I, Hunter V, Walker RI (1984) Synergistic effect of bacteroides, Clostridium, Fusobacterium, anaerobic cocci, and aerobic bacteria on mortality and induction of subcutaneous abscesses in mice. J Infect Dis 149:924-928.
- EbersoleJL, Feuille F, Kesavalu L, Holt SC (1997) Host modulation of tissue destruction caused by periodontopathogens: effects on a mixed microbial infection composed of Porphyromonasgingivalisand Fusobacteriumnucleatum. MicrobPathog 23:23-32.
- Araki H, Kuriyama T, Nakagawa K, Karasawa T (2004) The microbial synergy of Peptostreptococcus micros and Prevotellaintermediain a murine abscess model. Oral MicrobiolImmunol 19:177-181.
- Citron DM, PoxtonIR, Baron EJ (2007) Bacteroides, Porhyromonas, Prevotella, Fusobacterium, and other anaerobic Gram-negative rods. In: Murray PR, Baron EJ, Jorgensen JH, Landry ML, Pfaller MA, editors. Manual of clinical microbiology, 9th edition. Washington DC: American Society for Microbiology. pp.911-932.
- Kuriyama T, Karasawa T, Nakagawa K, Yamamoto E, Nakamura S (2001) Incidence of ß-lactamase production and antimicrobial susceptibility of anaerobic gram-negative rods isolated from pus specimens of orofacialodontogenic infections. Oral MicrobiolImmunol 16: 10-15.
- Kuriyama T, Karasawa T, Nakagawa K, Kawashiri S, Nakanishi I, et al (2000) Characterization of bacterial orofacial infections using a new murine model. MicrobPathog 29:115-120.
- Relman DA, Falkow S (2005) A molecular perspective of microbial pathogenicity. In: Mandell GL, Bennett JE, Dolin R, editors. Principles and practice of infectious diseases 6th edition. Philadelphia: Churchill Livingstone. pp.3-14
- Pérez-ChaparroPJ, Gracieux P, Lafaurie GI, DonnioPY, Bonnaure-Mallet M (2008) Genotypic characterization of Porphyromonasgingivalisisolated from subgingival plaque and blood sample in positive bacteremia subjects with periodontitis. J ClinPeriodontol 35:748-753.
- Wang X, Maegawa T, Karasawa T, Kozaki S, Tsukamoto K, et al (2000) Genetic analysis of type E botulinumtoxin-producing Clostridium butyricumstrains. Appl Environ Microbiol 66: 4992-4997.
- National Committee for Clinical Laboratory Standards (1997) Methods for antimicrobial susceptibility testing of anaerobic bacteria. 4th edition.Approved standard.Document M11-A4. Villanova: NCCLS.
- Khemaleelakul S, Baumgartner JC, Pruksakorn S (2002) Identification of bacteria in acute endodontic infections and their antimicrobial susceptibility. Oral Surg Oral Med Oral Pathol Oral RadiolEndod 94:746-755.
- SiqueiraJFJr, Rôças IN, Souto R, Uzeda M, Colombo AP (2001) Microbiological evaluation of acute periradicular abscesses by DNA-DNA hybridization. Oral Surg Oral Med Oral Pathol Oral RadiolEndod 92:451-457.
- Riggio MP, Aga H, Murray CA, Jackson MS, Lennon A, et al (2007) Identification of bacteria associated with spreading odontogenic infections by 16SrRNA gene sequencing. Oral Surg Oral Med Oral Pathol Oral RadiolEndod 103:610-617.
- Baumgartner JC, SiqueiraJFJr, Xia T, Róças IN (2004) Geographical differences in bacteria detected in endodontic infections using polymerase chain reaction. J Endod 30:141-144.
- Sakamoto M, Rôças IN, SiqueiraJFJr, Benno Y (2006) Molecular analysis of bacteria in asymptomatic and symptomatic endodontic infections. Oral MicrobiolImmunol 21:112- 122.
- Roberts SA, Shore KP, Paviour SD, Holland D, Morris AJ (2006) Antimicrobial susceptibility of anaerobic bacteria in New Zealand: 1999-2003. J AntimicrobChemother 57:992-998.
- Fosse T, Madinier I, Hitzig C, Charbit Y (1999) Prevalence of ß-lactamase-producing strains among 149 anaerobic gram-negative rods isolated from periodontal pockets. Oral MicrobiolImmunol 14:352-357.
- Kuriyama T, Karasawa T, Williams DW, Nakagawa K, Yamamoto E (2006) An increased prevalence of ß-lactamase-positive isolates in Japanese patients with dentoalveolar infection. J AntimicrobChemother 58:708-709.
- Carapetis J, Gardiner D, Currie B, Mathews JD (1995) Multiple strains of Streptococcus pyogenesin skin sores of aboriginal Australians. J ClinMicrobiol 33: 1471-1472.
- Fukui K, Kato N, Kato H, Watanabe K, Tatematsu N (1999) Incidence of Prevotellaintermediaand Prevotellanigrescenscarriage among family members with subclinical periodontal disease. J ClinMicrobiol 37: 3141-3145.
- Suchett-Kaye G, Decoret D, Barsotti O (1998) Clonal analysis by ribotyping of Fusobacteriumnucleatumisolates obtained from healthy young adults with optimal plaque control. J Periodontal Res 33:179-186.
- Marsh PD, Martin MV, Lewis MAO, Williams DW (2009) Oral microbiology. 4th edition. Edinburgh: Churchill Livingstone Elsevier. pp.103-145.
- Yanagisawa M, Kuriyama T, Williams DW, Nakagawa K, Karasawa T (2006) Proteinase activity of Prevotellaspecies associated with oral purulent infection. CurrMicrobiol 52:375-378.
- Amano A, Nakagawa I, Okahashi N, Hamada N (2004) Variations of Porphyromonasgingivalisfimbriae in relation to microbial pathogenesis. J Periodontal Res 39:136-142.









