Jafari-Sales A1,2*, Sayyahi J1, Akbari-Layeg F3, Mizabi-Asl M4, Rasi-Bonab F5, Abdoli-senejani M2 and Ezdiyadi M5
1Ahar Branch, Islamic Azad University, Ahar, Iran
2Department of Microbiology, Kazeroon branch, Islamic Azad University, Kazeroon, Iran
3Department of Microbiology, Urmia Branch, Islamic Azad University, Urmia, Iran
4University of Alzahra, Tehran, Iran
5Marand Branch, Islamic Azad University, Marand, Iran
*Corresponding Author:
Jafari-sales A
Department of Microbiology, Kazeroon branch
Islamic Azad University, Kazeroon, Iran.
Tel: +98(0) 9147611841
Fax: +98(0) 4142274746
E-mail: A.jafari_1392@yahoo.com
Received date: August 17, 2017; Accepted date: September 08, 2017; Published date: September 12, 2017
Citation: Jafari-Sales A, Sayyahi J, Akbari-Layeg F, Mizabi-Asl M, Rasi-Bonab F, et al. (2017) Identification of gyrA Gene in Ciprofloxacin-Resistant Enterococcus Faecalis in Strains Isolated from Clinical Specimens in Hospitals and Clinics of Tabriz and Marand Cities. Arch Clin Microbiol. Vol. 8 No. 5:63. doi: 10.4172/1989-8436.100063
Copyright: © 2017 Jafari-Sales A, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Keywords
Enterococcus faecalis; Ciprofloxacin; gyrA
Introduction
Enterococci are gram-positive and catalase-negative cocci. They can grow at the presence of 6.5% salt and 40% bile salts. These bacteria are considered natural intestinal flora of warm blooded animals, including humans. The most common enterococci involved in human infections are Enterococcus faecalis (85-90%) and faecium (5-10%) [1]. Hospital infections lead into prolonged hospitalization, and thereby they cause significant increase in health care costs of the country [2]. Enterococcus is considered as the important cause of hospital infections and the infections acquired from the community In the last two decades [3,4]. Enterococci have been identified as one of the most important human pathogens. These organisms are inherently resistant to a large number of antibiotics. Moreover, they can increasingly obtain new resistances. Enterococcus faecalis, as the most common species, accounts for more than 80% of infections caused by enterococci such as endocarditis, bacteremia, urinary tract, hospital, infants and central nervous system infections [5]. Increasing role of enterococci in hospital infections can be due to the increased impact of selective pressure resulting from the undue use of various antibiotics. One of the drugs that is used to treat Enterococcal infections, especially urinary tract infections, is fluoroquinolone antibiotics. The common of them is ciprofloxacin. As antibiotics are used without restriction in Iran, and as Enterococcus faecalis easily gains resistance genes, strains of this bacterium have indicated varying and increased degrees of resistance to ciprofloxacin. As ciprofloxacin used widely, resistance to it among clinical specimens has caused problems in treatment of Enterococcal infections. Controlling Enterococcus faecalis infections is very difficult, especially its multi-resistance types, and there is no definite treatment for it. Hence, controlling the transmission and prevalence of these microorganisms is very important [6]. There are effective mechanisms for the acquisition and transfer of resistant genes in a number of these species and other bacterial species. Gentamicin-resistant encoding genes are able to displace between Enterococcus faecalis strains with a high frequency, through pheromone-responsive plasmids. The occurrence of this phenomenon on Iranian strains has already been confirmed [7]. While several studies have been conducted on virulence factors of these bacteria, among different strains isolated from healthy and diseased people, no virulence factor has been found so far, which suggests pathogenicity can be attributed to them. The most common causes of human diseases by this group of bacteria are the two types of Enterococcus faecalis and Enterococcus faecium [8]. While no protein secretion toxin has been identified in enterococci so far, their pathogenicity probably occurs due to activity of a set of factors involved in pathogenesis, such as proteins or carbohydrates involved in binding of the bacteria to the gut and genital ulcer epithelium, and cumulative factor involved in exchange of plasmid and other factors [9]. Combination of ciprofloxacin and metronidazole is one of these medicinal compounds. Ciprofloxacin is the generic international name for a kind of synthetic antibiotic produced by Bayer Pharmaceuticals under brand names of Cipro and Ciproxin. This antibiotic belongs to the group of fluoroquinolones and it is a bactericide. Its mechanism of action is to prevent bacterial DNA replication by binding to the DNA giraze enzyme [10]. The current research was conducted to identify ciprofloxacin-resistance gene in Enterococcus faecalis.
Methodology
Collection and identification of samples
In total, 196 different clinical samples including urine and blood were collected from hospitalized patients and outpatients of Tabriz and Marand hospitals and clinics during seven months since October 2014 to April 2015. Samples were detected in terms of genus, through gram staining, by tests of catalase, sensitivity to bacitracin, growth on a salt environment of 6.5% sodium salt and hydrolysis test of alpha-pyrrolidonyl-beta-naphthylamide (PYR) (all culture mediums were purchased from Merck Company) [11,12]. Polymerase chain reaction (PCR) molecular identification method was used for final determination of genus and species.
Identification of genus and species based on ISR gene
The sequence of ISR section belonging to all Enterococcal species, including Enterococcus faecalis, was extracted from NCBI databank, and was aligned by Clustal x bioinformatics software and was designed through the specific section of faecalis primer species by OLIGO7 program, so that these primers can reproduce ISR fragment only in the polymerase chain reaction. The design primers are shown in Table 1.
| Primer |
Sequence |
| EISRF |
5?- CTAAGGAATATTACGGAAAT-3? |
| EISRR |
5?- TCTAGCGATAGAAGGTTA-3? |
Table 1: Primers used in PCR reaction.
The following steps were taken to extract the genomic DNA from Enterococcus bacteria strains
The bacterial specimens were incubated on liquid culture medium at 37°C for 24 hours. Ten milliliters of bacterial cultures were centrifuged for 10 minutes at 7000 g, and the supernatant was discarded and plates were used to extract the DNA. Firstly, some liquid nitrogen was poured onto the plates to facilitate the smooth transferring of plates into porcelain mortar. Then, plates were transferred to the porcelain mortar. Adding a little excess liquid nitrogen and using pounder of the mortar, a powder was provided that had single bacterial cells. Lysing buffer, shown in the following table, was used to lyse the cells. After pouring 700 μl of lysing buffer in sterile porcelain mortars that contained powdered cells, the cells were crushed and transferred to 1.5 ml vials. The vials were placed in Ben Mary at 60°C for one hour. 1.4 μl RNAase was added to it and it was placed in Ben Mary for 30 minutes at 37 degrees centigrade. After 30 minutes, the vials containing lysed cells were centrifuged for 5 minutes at 12000 g and the supernatant was transferred to other vials and at an equal volume, chloroform-isoamyl alcohol was added to that it was shaken gently at a ratio of 1:24. Then, it was centrifuged in the model centrifuge for 10 minutes at a rate of 12,000. The supernatant was removed and an equal volume of cold isopropanol was added and was stored at -20° C for one night and again was centrifuged in the model centrifuge for 10 minutes at a rate of 12,000. The supernatant was discarded and DNA was dried by placing the vials at room temperature. Finally, dried DNA was dissolved in 50 μl of deionized water. In order to find the quality and quantity of DNA extraction, 5 μl of the DNA dissolved in 50 μl of deionized water was electrophoresed by using 1% agarose gel. Polymerase chain reaction in 25 μl volume was carried out with the components of table 2 using the designed primers.
| 2x Master Mix |
12.5 µl |
| dH2O |
9.5 µl |
| Primer forward |
1 µl |
| Primer Revers |
1 µl |
| DNA |
1 µl |
Table 2: Concentration of the components forming polymerase chain reaction.
One percentage agarose gel was used to isolate PCR product, so that the amplified specimens were mixed with the loading buffer at the ratio of 6 to 1 and were loaded onto gel wells. After that electrophoresis was completed, the considered gel was observed through staining and the amplified DNA strips in all samples as single strip and with a size of approximately 300 pairs of nucleotides and with appropriate concentrations using UV light, images were taken. After analyzing the obtained samples, two samples of PCR products with four different sequences for sequencing by Fazapajooh Company of Tehran were sent to Korean Macrogan Company in order to ensure that the obtained coccus bacteria really belong to Enterococcus faecalis, and the result was identified after blast of Enterococcus faecalis.
Antibiotic sensitivity test
In order to examine the drug-resistance to using the standard method of Kirby Baur, resistance of Enterococcus faecalis strains to ciprofloxacin 5 mg antibiotic, prepared from Padtan Teb Co, was examined on Muller Hinton Agar (Merck) environment [13]; the minimum inhibitor concentration of MIC was determined through E-Test method (HighMedia Indian Company) based on the Clinical Standards Institute (CLSI) and the findings were interpreted according to CLSI standards guideline [14].
Gene identification (gyrA)
In order to detect resistance of genes by DNA method, gyrA resistance gene was examined. Sequence of primers is listed in Table 3. The thermo-cycle of Thermocycler machine for proliferation of gyrA resistance genes of 35 cycles was as follows:
| Product size (bp) |
Sequence of primer |
Gene |
| |
GCAATGAGTGTTATCGT
TCTGGTCCAGGTAACACTTCC |
gyrA |
Table 3: Sequence of the primers used for ciprofloxacin-resistance genes.
gyrA: Initial denaturation, 94°C, 3', Denaturation, 94°C, 60'', Annealing, 58°C, 60'', Extension, 72°C, 60''.
Results
After that polymerase chain reaction (PCR) was carried out, it was found that among 196 tested Enterococcus samples, only 194 samples (98.97%) are Enterococcus in terms of genotype test, and among these, only 93 samples belonged to faecalis species Figure 1.
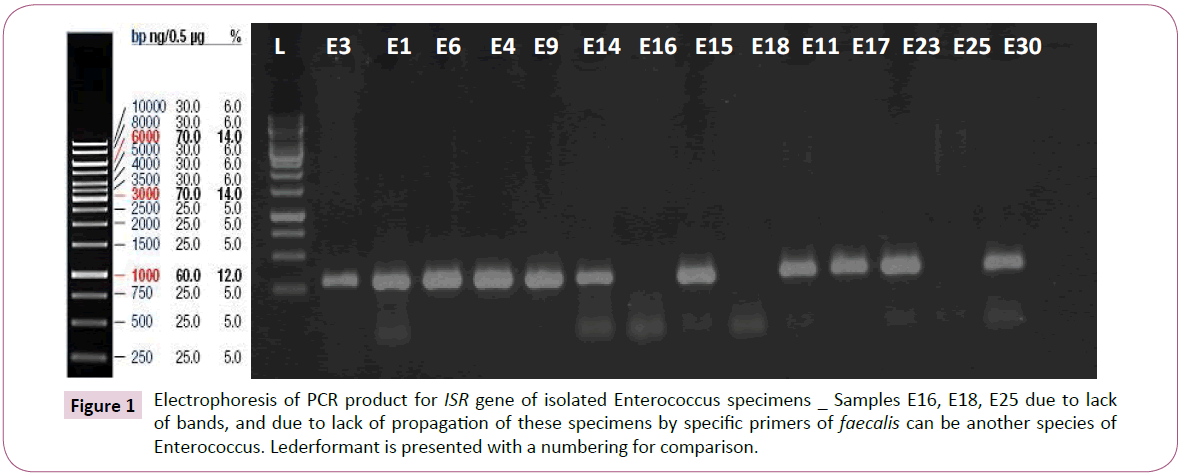
Figure 1: Electrophoresis of PCR product for ISR gene of isolated Enterococcus specimens _ Samples E16, E18, E25 due to lack of bands, and due to lack of propagation of these specimens by specific primers of faecalis can be another species of Enterococcus. Lederformant is presented with a numbering for comparison.
Two samples of E15 and E3 were sent to Macrogene Co. for sequencing. They also were diagnosed using blast in NCBI, with E3 faecalis sample, but E15 sample showed many mutations in this gene, that had not been reported so far with any other bacteria. Figure 2 illustrates the sequencing sample that has been sequenced with a very good quality. In addition, findings of blast of two cases in ncbi are illustrated in Figure 2. Findings revealed that this strain had a percentage of similarity and nucleotide of overlapping region with Enterococcus. The overlapping region is shown as Plus/Plus in the Figure. New mutation in E15 bacteria is recorded in NCBI databank.
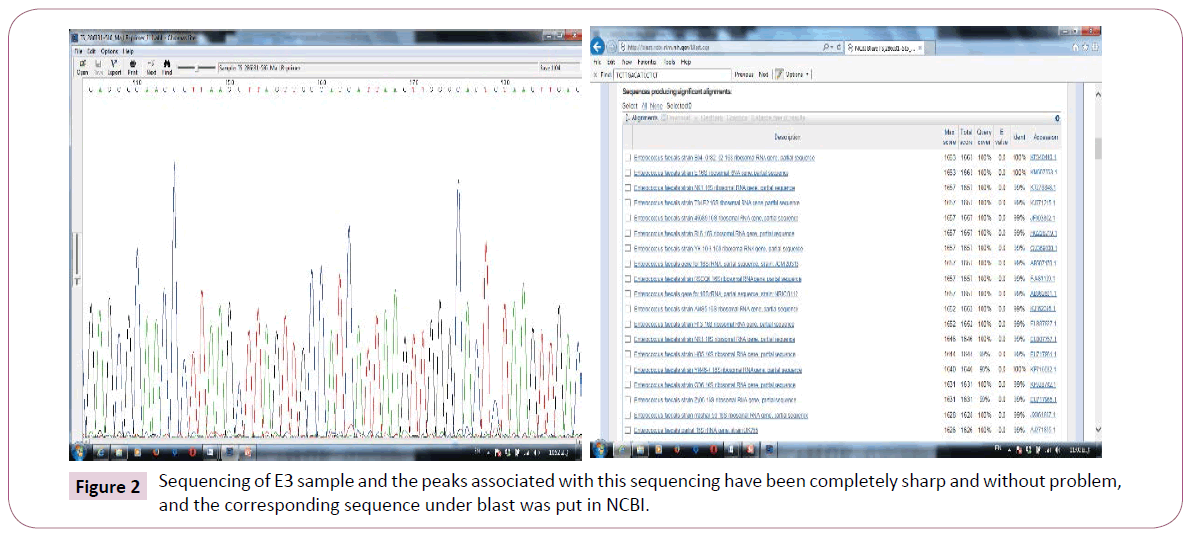
Figure 2: Sequencing of E3 sample and the peaks associated with this sequencing have been completely sharp and without problem, and the corresponding sequence under blast was put in NCBI.
However, in the case of E15, such a similarity is not seen with any other bacteria in the world, and a similarity of 93% with faecalis species means that it has the highest similarity with this species and it has been able to reproduce with the specific ISR primers too in order to be selected for subsequent studies and sequencing. In Figure 3, two samples of the relevant mutations and peaks are presented, that it can be said that this species is likely to be reported in Iranian hospitals.
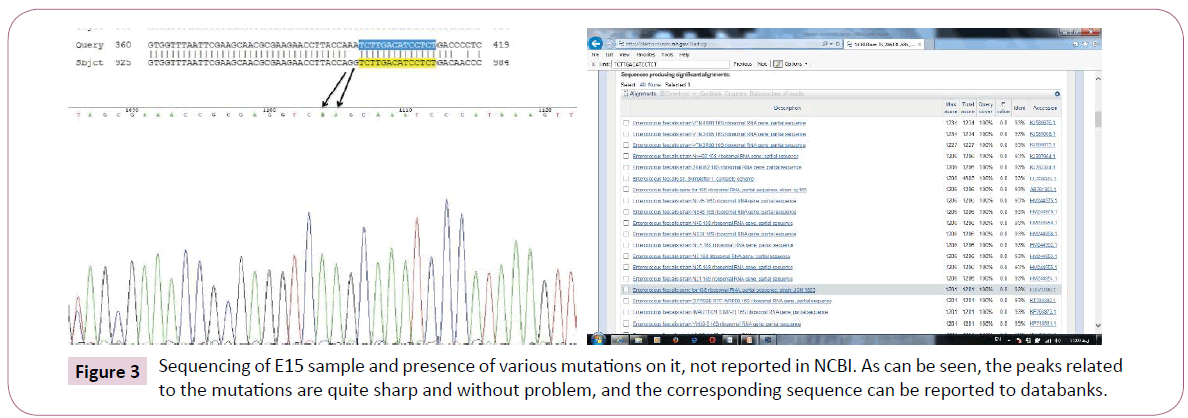
Figure 3: Sequencing of E15 sample and presence of various mutations on it, not reported in NCBI. As can be seen, the peaks related to the mutations are quite sharp and without problem, and the corresponding sequence can be reported to databanks.
According to antibiotic sensitivity test, Enterococcus faecalis species showed the highest resistance to tetracycline and ciprofloxacin antibiotics and the most sensitivity to antibiotics Tables 3 and 4.
| Antibiotic |
S |
I |
R |
| Vancomycin |
72.6 |
5.3 |
22.1 |
| Ampicillin |
66.31 |
2.09 |
31.6 |
| Linezolid |
89.4 |
3.2 |
7.4 |
| Gentamicin |
50.6 |
10.5 |
38.9 |
| Teicoplanin |
77.9 |
0 |
22.1 |
| Ciprofloxacin |
40 |
18.9 |
41.1 |
| Tetracyclines |
54.7 |
2.1 |
43.2 |
| Dalfopristin |
60 |
10.5 |
29.5 |
S: Sensitive; I:Intermediate; R: Resistant
Table 4: Distribution of percentage of resistance and sensitivity of Enterococcus strains to antibiotics.
Determining the minimum inhibitory concentration in the present study revealed that 40 samples were resistant to ciprofloxacin. Among resistant strains, 10 strains had a minimum inhibitory concentration of 4 μg/ml and 15 strains had a minimum inhibitory concentration of 12 μg/ml and 18 strains had a MIC value greater than 32 μg/ml Figure 4. Ciprofloxacin-resistant strains have been isolated from urine specimens with a frequency of 29 specimens and wound specimens with a frequency of 11 specimens. In this study, 21 strains resistant to Vancomycin were isolated from the outpatient section and 19 from the admission section.

Figure 4: Determination of the minimum inhibitor concentration (MIC) by E-Test.
Among 40 ciprofloxacin-resistant specimens, 92.5% contained gyrA gene Figure 5.
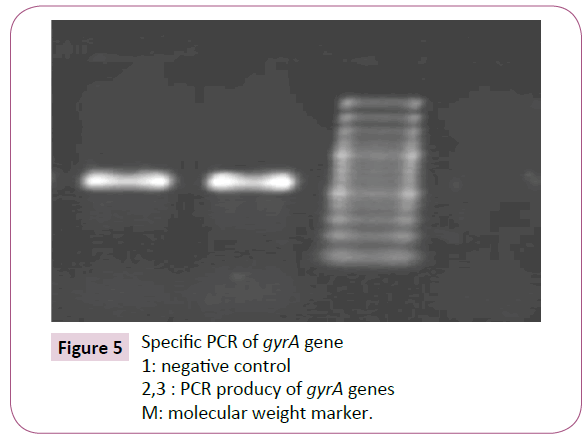
Figure 5: Specific PCR of gyrA gene
1: negative control
2,3 : PCR producy of gyrA genes
M: molecular weight marker.
Discussion
Ciprofloxacin resistance to enterococci was not common until 1985-1896, but resistance significantly increased to 15.2% in 1990. Resistance among isolated faecalis strains to ciprofloxacin in the next years reached to 31.5% [15]. Based on the research conducted by saifi in 2008, high resistance (approximately 40%) to ciprofloxacin was observed among clinical isolates and sewage, which can be due to using this drug in treatment of various types of urinary tract infections as the first selected option [13]. As enterococci are the normal flora of human and warm-blooded animals’ intestines, they can easily enter to environment through feces and are abundantly isolated in the outside environment from raw food, vegetables, surface water and sewage. Enterococci-resistant strains can also enter the normal flora of humans’ intestine in this transfer chain [16]. Increased role of enterococci in hospital infections can be due to the use of antibiotics that people are resistant to them [15,16]. One of the most common drugs used to treat Enterococcal infections is fluoroquinolone antibiotics is ciprofloxacin, which is especially prescribed in Enterococcal urinary tract infections. As using this antibiotic in used without restriction in Iran, Enterococcus faecalis easily gains resistance genes. The bacterium has showed high levels of resistance to ciprofloxacin, led to difficulty in controlling the treatment and prevalence of Enterococcal infections. MIC values above 4 μg/ml of ciprofloxacin in Enterococcus faecalis are considered as resistant [17]. Iran is a country where antibiotic is used without restriction on one hand, and antibiotics are used inappropriately and inadequately on the other hand. Causing drug resistance, the intestine is colonized with Enterococcal resistant strains, and this resistance is also transferred to sensitive strains. These resistant strains in feces results in sewage contamination and dissemination of resistance genes. If people colonized with these strains are hospitalized for any reason, resistant strains might be transferred to other patients. It might be transferred through the contaminated hands of the hospital staff or contaminated equipment. When the hospitalization period is longer, the rotation of these resistant strains in all parts of the hospitals will be more risky; and finally, after discharge of these patients, these strains might release to community. Existence of resistant hospital and outpatient isolates within a clone is an alarm that should be taken into consideration seriously by the health care system. Common clones that have previously been restricted to hospitals have been transferred from hospitals to the society due to various reasons and lack of proper health care. This problem is very dangerous due to rotation of resistant strains in the community. Preventive protocols must be implemented in hospitals in order to prevent transmission of resistant clones to the community and their rotation; otherwise, we will encounter a wide range of treatment-resistant Enterococcal infections due to increased resistant strains in the community. In addition, due to transfer of resistance genes to similar or non-similar strains and other bacteria genus, there will be a potential risk of creation of other bacterial infections. Colonization and infection with enterococci have been growing significantly. In such a situation, infection control is very complicated and treatment of such infections is very important. In this situation, control of transmission of such kinds of microorganisms is very important and genotyping methods are very effective in detecting colonality of strains and controlling them better. The health care system of each society should identify the important and common factors of the hospital pathogens appropriately and determine the exact pattern of their antibiotic resistance so that effective prevention and treatment methods can be used in order to control them. In this research, like the research conducted by Feizabadi et al. most of the Enterococcal strains were isolated from urine specimens. This suggests the importance of establishment of enterococci in urinary tracts. Enterococci are the primary cause of infection among gram positive infectious cocci in urinary tracts and the third most common cause of bacterial infection in women urinary tracts in Iran after Escherichia coli and Klebsiella pneumoniae [7]. Fathollahzadeh et al. in 2006 investigated people with urinary tract infection in three hospitals in Tehran. They reported that the prevalence of Enterococcal species is 57% Enterococcus faecalis, 30% Enterococcus faecium, and 13% other species. These results are in line with results of the current study [18]. In Enterococcus faecalis, mutational faults in the genes gyrA and gyrB are the main causes of high resistance to ciprofloxacin (DNA gyrase consists of two subunits, GyrA and GyrB, encoded by the gyrA and gyrB genes, respectively) [19-20]. Based on the research conducted by Seyfi in 2013, 50 strains of Enterococcus faecalis were resistant to ciprofloxacin, of which 98% were positive in terms of gyrA gene. It is also consistent with the findings of the current study [8]. In a study conducted by Torell et al in 2003, 17% of ciprofloxacin-resistant strains contained gyrA gene [4].
Conclusion
Ciprofloxacin-resistant Enterococcus faecalis strains have diverse colonality. As high values of MIC are seen in hospital and outpatient specimens, it can be stated that they do not have a uniform distribution, and this suggests the transfer of resistance genes at high levels in the Enterococcal population. In fact, individuals colonized by these strains are considered reservoirs for dissemination of resistance gene in the community, which requires more caring programs.
20621
References
- Saeed AK, Mohamad SN, Ashraf AK (2005) Selective isolation of multi drug resistant Entrococcus spp, from poultry and dairy farms: detection of virulence and vancomycin resistance gene markers by PCR. Molecular Cellular Probes 19: 27-34.
- Dar-odeh NR, Abu Hammad O, Shehabi AA (2008) Prevalence of putative virulence factor and antimicrobial susceptibility of Enterococcus faecalis isolates from patients with dental diseases. BMC Oral Health 1: 8-17.
- Goossens H, Jabes D. Rossi R, Lammens C, Privitera G, et al. (2003) European survey of vancomycin-resistant enterococci in at-risk hospital wards and in vitro susceptibility testing of ramoplanin against these isolates. J Antimicrob Chemother 3: 5-12.
- Torell E, Kuhn I, Olsson-Liljequist B, Haeggman S, Hoffman BM, et al. (2003) Clonality among ampicillin-resistant Enterococcus faecium isolates and relationship with ciprofloxacin resistance. J Clin Microbiol. Infect 9: 1011-1019.
- Huycke MM, Sahm DF, Gilmore MS (1998) Multiple drug resistant Enterococci: the nature of the problem and an agenda for future. Emerg Infect Dis 4: 239-249.
- Gordillo ME, Sing KV, Murray BE (1993) Comparison of Ribotyping and Pulsed- Field Gel Electrophoresis for subspecies differentiation of strains of Enterococcus faecalis. J Clin Microbiol 31: 1570-1574.
- Feizabadi MM, Asadi S, Khatibi S, Etemadi G, Parvin M, et al. (2004) A survey on the resistance pattern of Enterococcus faecalis and Enterococcus faecium strains in Labfinejad and Shahid Chamran hospitals during the years 2000-2003. pajoohande 9: 9-15.
- Moinian M, Pourshafie MR, Eidi A, Safarpour E, saifi M (2013) Genetic Diversity of Ciprofloxacin Resistant Enterococcus Fecalis Strains Isolated from Clinical Samples by Field Electrophoresis Field. IJIDTM Journal 18: 21-28.
- Flemmig TF, Milian E, Karch H, Klaiber B (1998) Differential clinical treatment outcome after systemic Metronidazole and amoxicillin in patients harboring Actinobacillus Actinomycetemcomitans and/or Prophyromonas Gingivalis. J Clin Periodontol 25: 380-387.
- Leavis, HL, Willems RJ, Bonten MJ (2006) High-level ciprofloxacin resistance from point mutations in gyrA and parC confined to global hospital-adapted clonal lineage CC17 of Enterococcus faecium. J Clinl Microbiol 44: 1059-1064.
- Facklam RR, Collins MD (1989) Identification of Enterococcus species isolated from human infections by a conventional test scheme. J Clin Microbiol 27: 731-734.
- Saunders GL, Bodonaik NC (2006) Resistance in clinical isolates of Enterococcus faecalis encountered at the university hospital of the West Indies, Jamaica. West Indian Med J 55: 194-196.
- Saifi M, Soltan MM, Pourshafie MR, Eshraghian MR, Salari MH, Shirazi MH (2008) High-level resistance of E. faecium and E. faecalis isolates from municipal sewage treatment plants to gentamycin. Iranian J Pub Health. 37: 103-107.
- Modi GB, Soni ST, Patel KJ, Goswami HM, Vegad MM (2012) Prevalence of vancomycin resistant Enterococci in tertiary care hospital, western, India. Int J Microbiol Res 4: 182-185.
- Schaberg DR, Dillon W, Terpenning MS, Robinson KA, Bardley SF (1992) Increasing resistance of Enterococci to ciprofloxacin. Antimicrob Agents Chemother 36: 2533- 2535.
- Tankovic J, Mahjoubi F, Courvalin P, Duval J, Leclerco R (1996) Development of fluoroquinolone resistance Enterococcus faecalis and role of mutations in the DNA Gyrase gyrA gene. Antimicrobial Agents Chemother 40: 2558-2561.
- Manero A, Blanch AR (1999) Identification of Enterococcus spp. with a biochemical key. Appl. Environ. Microbiol 65: 4425-4430.
- Fatholazadeh B, Hashemi BF, Emaneini M, Aligholi M, Nakhjavani AF, et al. (2006) Detection of vancomycin resistant Enterococci (VRE) isolated from urinary tract (UTI) in Tehran, Iran. DARU 14: 141-145.
- Onodera Y, Okuda J, Tanaka M, Sato K (2002) Inhibitory activities of quinolones against DNA Gyase and Topoisomerase IV of Enterococcus faecalis. Antimicrob Agents Chemother 46: 1800-1804.
- Korten V, Huang W, Murray B (1994) Analysis by PCR and Direct DNA Sequencing of gyrA mutations associated with fluoroquinolone resistance in Enterococcus faecalis. Antimicrobial Agents and Chemotherapy 38: 2091-2094.










