Introduction
Polycythemia vera (PV) along with essential thrombocythemia (ET), idiopathic myelofibrosis (IMF), chronic myelocytic leukemia (CML) and essential thrombocythemia (ET) are important myeloproliferative disorders (MPD) that are demonstrated to arise clonally from a pluripotent hematopoietic stem cell (Fialkow et al., 1967; Adamson et al., 1976). It is a well known stem cell anomaly more specifically identified as a panhyperplastic, malignant and neoplastic marrow disorder. The disease causes high level of absolute red blood cell mass as well as increased white blood cell (myeloid) and platelet (megakaryocytic) production. (Berlin, 1975; Landolfi, 1998; Streiff et al., 2002). A constant hallmark of PV as well as other MPD bone marrow cells is their hypersensitivity to several cytokines (Prchal and Axelrad, 1974; Dai et al., 1992; Correa et al., 1994; Dai et al., 1994) and their ability to generate EPO-independent erythroid colonies in vitro (Prchal and Axelrad, 1974), commonly referred to as endogenous erythroid colonies (EECs).
The gene JAK2, codes for a tyrosine kinase and is essential for effective signaling in response to several cytokines (Parganas et al., 1998). Within this gene, there exists a hotspot at nucleotide position 1887 where a G is substituted by T. This leads to alteration of amino acid at position 617 of the protein from valine to phenylalanine. The mutation is commonly known as the JAK2 V617F. It has been found in a majority of patients suffering from PV and in some patients with ET and IMF (Baxter et al., 2005; James et al., 2005; Kralovics et al., 2005; Levine et al., 2005; Zhao et al., 2005).
The V617F mutation occurs within the negative regulatory domain thereby enhancing the JAK2 kinase activity which in turn causes cytokine- independent growth of cell lines and cultured bone marrow cells. It has been demonstrated that mutant JAK2 transfected into murine bone marrow cells produces erythrocytosis and subsequent myelofibrosis in recipient animals (Wernig et al., 2006; Lacout et al., 2006) suggesting a causal role for the mutation.
Till date, nucleotide sequencing remains one of the most popular and convenient method of detecting JAK2 V617F mutation (Ma et al., 2009; Ohyashiki et al., 2009). The method assists in direct detection of the single nucleotide polymorphism (SNP) apart from providing additional information on neighboring regions also for further detection of other cryptic mutations unlike other methods such as those based on real time PCR (Rapado et al., 2008) that rely on specificity of probe hybridization and has potential for generating false negatives if cryptic mutation occurs close to the target but within the probe hybridization region.
In this study we report an improved set of oligonucletoide primers for PCR amplification of the exon 12 region of JAK2 gene harboring the V617F mutation followed by their use in generating high quality nucleotide sequence employing an automated genetic analyzer.
Material and Methods
Samples Collection and storage
The study group comprised of 15 patients diagnosed for polycythemia vera (PV) and one for chronic myeloid leukemia (CML). Blood sample / bone marrow was collected from all individuals included in this study in K2-EDTA vacutainer (Becton Dickinson, Sun Diego, Calif.) at SN Gene laboratories, Surat, Gujarat (India) and transported to the central processing laboratory at geneOmbio Technologies, Pune, Maharashtra, India within 48 hour of collection at ambient temperature.
Nucleic Acid Extraction
Genomic DNA was extracted by phenol/chloroform method after proteinase K digestion following standard techniques (Sambrook and Russels, 2001). The quality of DNA was checked by agarose gel electrophoresis (0.8%) and the quantity was determined using spectrophotometer reading at 260 nm wavelength.
Designing of oligonucleotide primers
The nucleotide sequence of Homo sapiens chromosome 9 genomic contig reference assemblies were accessed from the public domain (https:// www.ncbi.nlm.nih.gov; Gene bank accession number NT_008413.17). The hotspot for mutation at amino acid position 617 in the JAK2 gene was subsequently identified and oligonucleotide primers designed using the Primer 3 software (Rozen and Skaletsky, 1999).
In silico analysis was performed to confirm the specificity of the probes. Electronic PCR result indicated that the primer pair would generate a PCR amplicon of size 440 bp flanking the mutational hotspot.
PCR Amplification and automated capillary electrophoresis
PCR amplification was performed using an automated thermal cycler (GeneAmp PCR System 9700; Applied Biosystems, Foster City, Calif ) using a genAmp Total PCR kit (geneOmbio Technologies, Pune, India). The reaction comprised of 250 nanograms of genomic DNA, 10x PCR buffer, 1.5 millimolar MgCl2, 200 micromolar (each) deoxynucleotide triphosphates, 20 picomoles of each primer, JAK2F (5’- GGCAGTTGCAGGTCCATATAA– 3’) and JAK2R (5’- TTCATTGCTTTCCTTTTTCACA –3’) (Sigma Aldrich, India) and 1 unit of Taq DNA polymerase.
The thermal cycling condition was as follows: 5 min for 95°C (initial denaturation) followed by 35 cycles of 95°C for 30 seconds, 56°C for 30 seconds, and 72°C for 1 min followed by a final extension for 7 min at 72°C. The PCR amplicon was analyzed by electrophoresis on a 2% agarose gel spiked with Ethidium bromide (0.5 micrograms/ml) in 0.5X TBE buffer) and visualized under UV transilluminator (260 nm).
The PCR products were purified by using Genpure PCR product purification kit (geneOmbio Technologies, Pune, India) and sequenced with JAK2F and JAK2R primers respectively using the BigDye Terminator v3.1 Cycle Sequencing Kit and analyzed with an automated genetic analyzer (Model 3130, Applied Biosystems, USA).
Data Analysis
A consensus sequence was generated from the double strand nucleotide sequence data for each sample and then multiple sequence alignment were performed using clustal W software (Thompson et al., 1994) with reference sequence (Gene bank accession number NT_008413.17).
Bone marrow karyotyping
GTG banding study of the clinical samples was done as described by Gadhia et al., (2005). Twenty well spread metaphases were studied and documented for each patient prior to recording of the data.
Results and Discussion
The term “myeloproliferative disorders” (MPDs) was coined by Dr. William Damashek in the year 1951 to address overlapping features of polycythemia vera, essential thrombocythemia (ET), myelofibrosis and myeloid metaplasia (MMM) (Damashek, 1951). One of the recent findings is the association of a gene coding for tyrosine kinase and belonging to the Janus kinase (JAK) family. Four different JAKs are reported in mammals. These are JAK1, JAK2, JAK3 and TYK2 (Valentino and Pierre, 2006). The JAK and STAT (another set of signal transduction proteins) signaling pathways play a pivotal role in establishing MPD by transmitting signals through the cytokine receptors to activate intracellular signaling pathway (Kwaja, 2006). Due to the V617F mutation in JAK2 gene, valine at codon position 617 is substituted by phenylalanine. This results in loss of auto-inhibition property of the encoded protein thus leading to constitutive transmission of signals from the erythropoietin (EPO) receptor, the thrombopoietin (TPO) receptor and the granulocyte colony stimulating factor (G-CSF) receptor in haemapoietic cells with high efficiency (Kwaja, 2006). This phenomenon is the primary cause of myelo-proliferation associated with polycythemia vera.
The discovery of this well known V617F mutation within JAK2 gene and its relation to MPD in the year 2005 can be compared to that of bcr-abl fusion gene responsible for chronic myeloid leukemia (CML) in the year 1983 (Bartram et al., 1983).
The primary aim of our study was to design, optimize and validate an improved and robust set of oligonucleotide primers that are well suited for identifying the V617F mutation within human JAK2 gene using PCR and automated DNA sequencing technology using Sanger’s chain termination method (Sanger and Coulson, 1975). The primer pair was designed with an aim to generate specific PCR amplicon of size >250 bp and <500 bp in size for efficient thermal amplification.
Thermal amplification of a region within the JAK2 gene covering the mutation hotspot at nucleotide position 1849 generated a PCR amplicon of 480 bp size (Figure 1). No non-specific amplification was detected across all samples and further the primer concentration was found to be optimized such that no significant primer-dimer formation could be seen on an agarose gel.
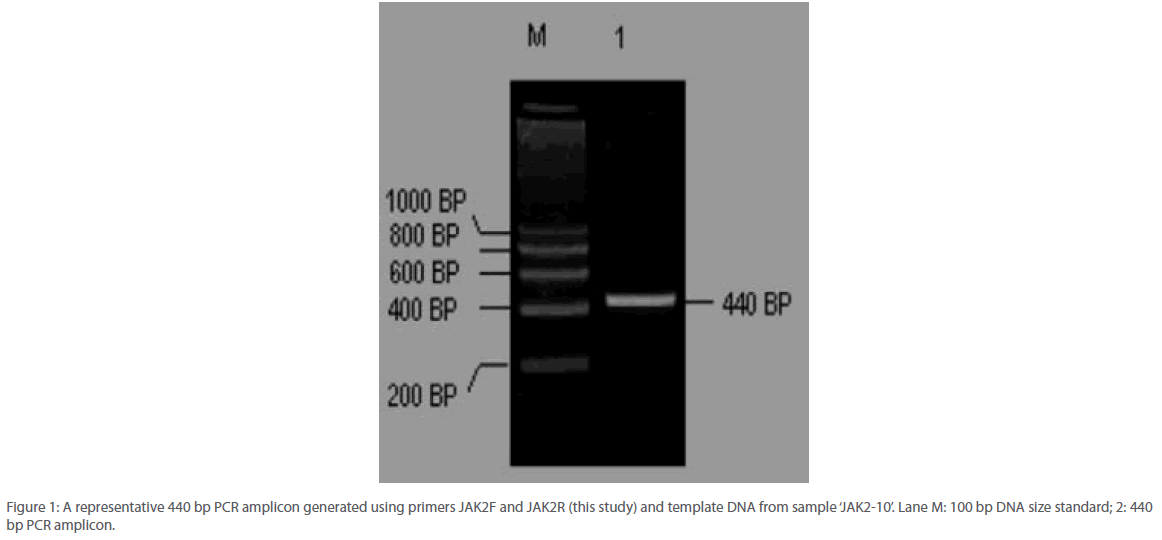
Figure 1: A representative 440 bp PCR amplicon generated using primers JAK2F and JAK2R (this study) and template DNA from sample ‘JAK2-10’. Lane M: 100 bp DNA size standard; 2: 440 bp PCR amplicon.
JAK2 1849G>T mutation was identified by two different sequencing reactions, each targeting one of the two strands of DNA from all the 105 patients analyzed. Example of the electropherograms generated is shown in Figure 2. Perfect correlation was found from the data generated from both the strands. Analysis revealed that the accuracy and reproducibility of the method was satisfactory (r2=0.99, p<0.0001). In order to obtain the linearity of the test developed, we undertook titration experiments where in DNA from a normal individual and that from a mutated one bearing 50% or 80% of the mutated JAK2 allele respectively were used. Mixture of DNA samples of varying ratio were amplified by conventional PCR and sequenced using the primers reported in this study. Liner relationship was observed between the content of patient DNA and amount of mutated allele that was detected by our method (r2=0.99). If was observed that the lower limit of detection was between 6-11% mutant allele. Detailed information of the oligonucleotide primers used for PCR and nucleotide sequencing in this study appear in Table 1.
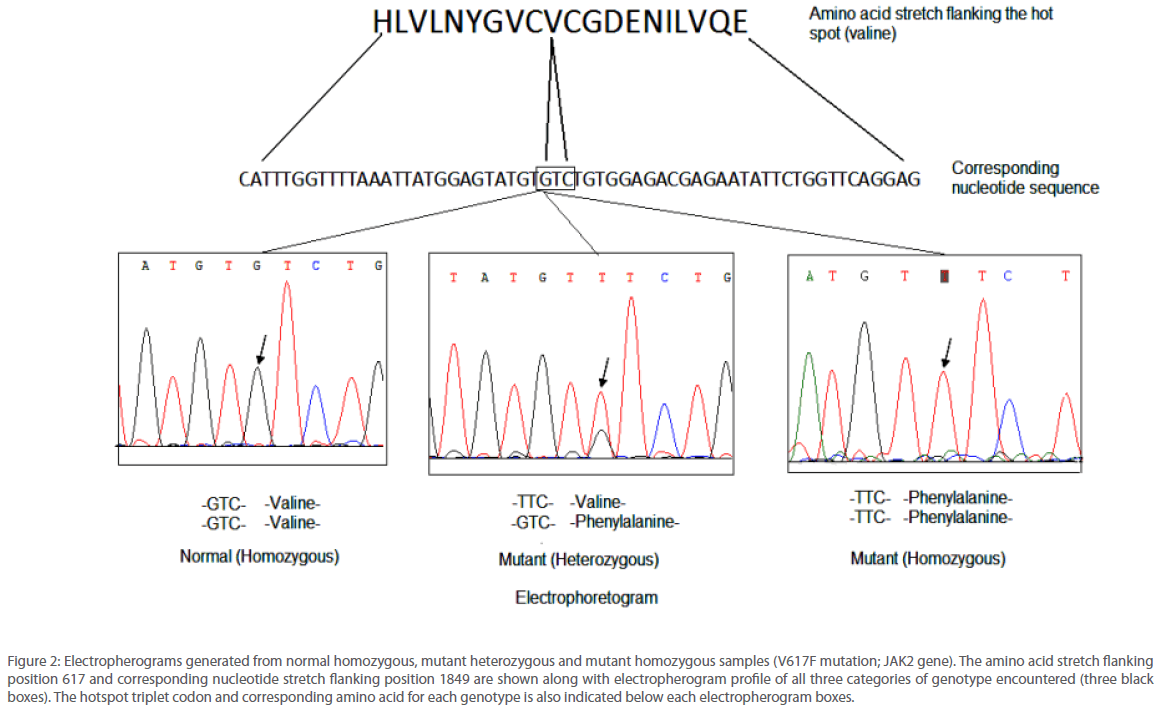
Figure 2: Electropherograms generated from normal homozygous, mutant heterozygous and mutant homozygous samples (V617F mutation; JAK2 gene). The amino acid stretch flanking position 617 and corresponding nucleotide stretch flanking position 1849 are shown along with electropherogram profile of all three categories of genotype encountered (three black boxes). The hotspot triplet codon and corresponding amino acid for each genotype is also indicated below each electropherogram boxes.
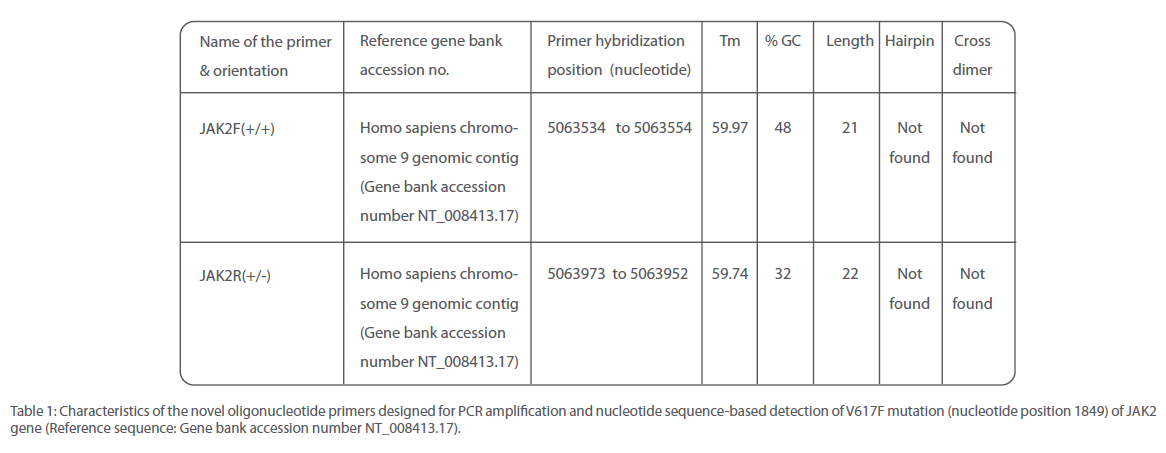
Table 1: Characteristics of the novel oligonucleotide primers designed for PCR amplification and nucleotide sequence-based detection of V617F mutation (nucleotide position 1849) of JAK2 gene (Reference sequence: Gene bank accession number NT_008413.17).
Out of 16 patients, one was found to be positive for Philadelphia chromosome and subsequently diagnosed with chronic myeloid leukemia (Figure 3). Remaining 15 patients were found to be suffering from Polycythemia vera. Out of them, 20% (3/15) were genotyped as homozygous mutant and 53.3% (8/15) as heterozygous mutant while 26.6% (4/15) were found to be of wild type with regard to V617F mutation within the JAK 2 gene (Figure 4). Almost 73.3% of the PV patients were found to carry the V617F mutation. This finding is in line with the report published by Jelinek et al., (2005) that also reported high proportion (86%) of PV patients to be positive for V617F mutation within the JAK2 gene. Table 2 summarizes the diagnosis report of all the 16 samples included in this study.
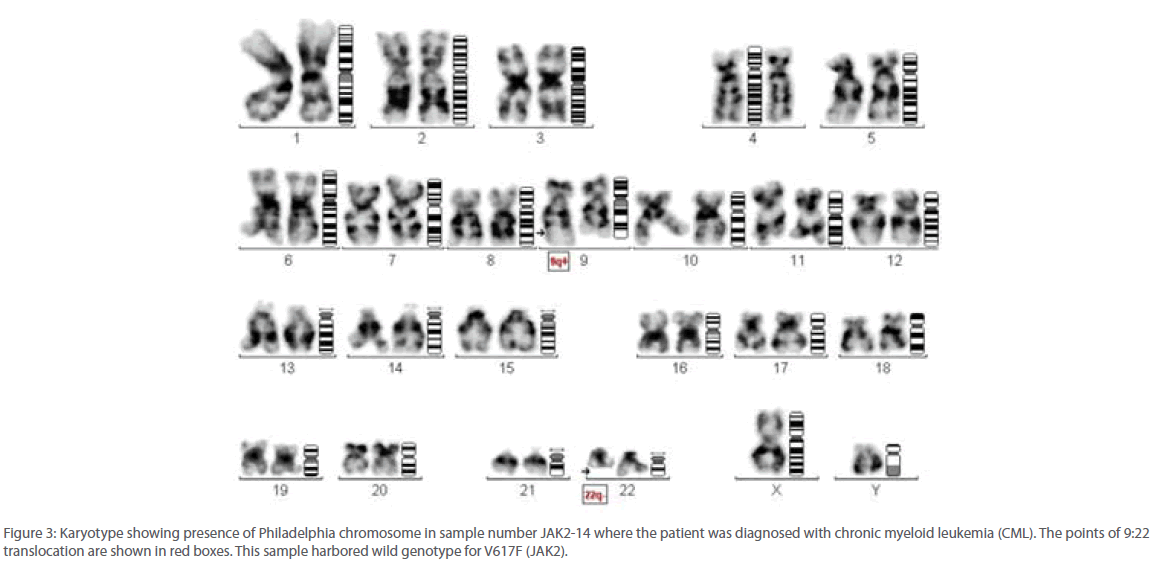
Figure 3: Karyotype showing presence of Philadelphia chromosome in sample number JAK2-14 where the patient was diagnosed with chronic myeloid leukemia (CML). The points of 9:22 translocation are shown in red boxes. This sample harbored wild genotype for V617F (JAK2).
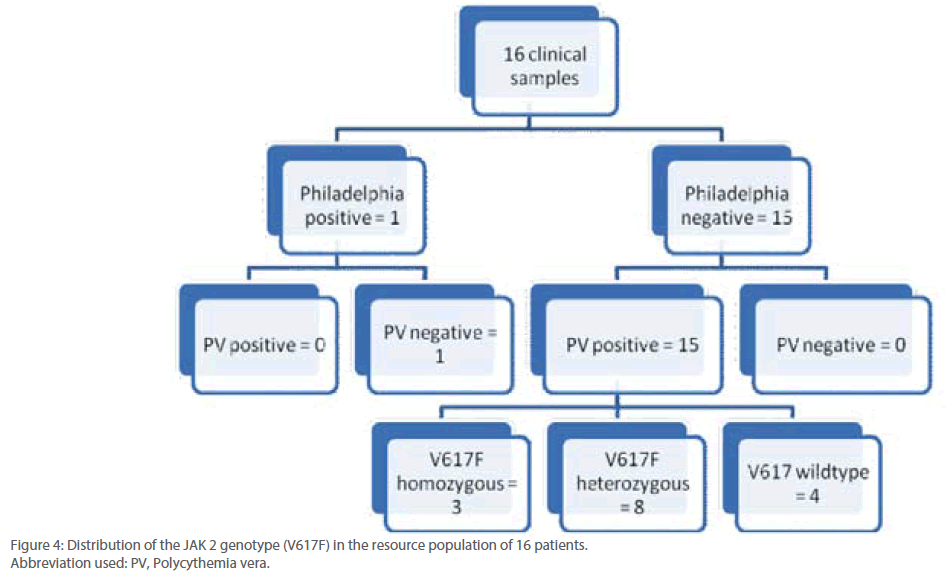
Figure 4: Distribution of the JAK 2 genotype (V617F) in the resource population of 16 patients.
Abbreviation used: PV, Polycythemia vera.
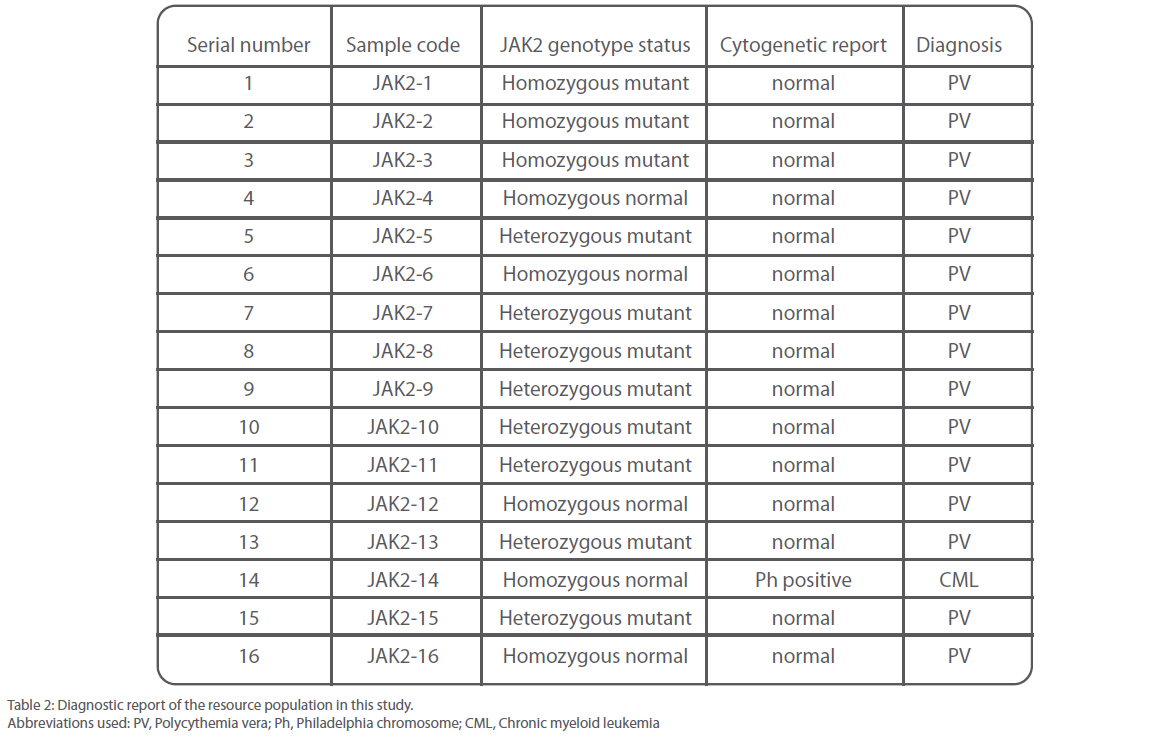
Table 2: Diagnostic report of the resource population in this study.
Abbreviations used: PV, Polycythemia vera; Ph, Philadelphia chromosome; CML, Chronic myeloid leukemia
The discovery of the drug named imatinib mesylate (IM) for the treatment of CML had its basis in the identification of bcr-abl fusion tyrosine kinase that had occurred almost two decades earlier (Agarwal, 2007). Therefore significant enthusiasm exist in the medical fraternity that following the discovery of JAK2 mutation there will be development of similar specific pharmacologic inhibitors of JAK2 with the potential to transform the treatment of PV, ET and MMM.
Superior molecular biology methods for error-free scanning of the mutational hotspot spanning nucleotide position 1849 within the JAK2 gene will have greater impact on the screening efficacy of patients. This study brings together a highly improved thermal amplification protocol for amplification of a region of the JAK2 gene coupled with an indigenously manufactured, low cost PCR product purification kit that in combination generated a pure and specific amplicon which formed the basis of generating good electropherograms. The oligonucleotide primers designed for this assay functioned with equal efficiency both as PCR as well as nucleotide sequencing primers and were capable of consistently generating high quality read of bases that is vital to accurate identification of heterozygotes.
Methods of identification of single nucleotide polymorphisms using nucleotide sequencing methods has the lone advantage of being capable of detecting the altering base directly rather than by indirect methods such as altering restricted PCR amplicon profile (RFLP) (Mukhopadhyaya et al., 2000) or using allele specific probes (Taqman chemistry based allelic discrimination or ARMS protocol) (Salvi et al., 2004; McWeeneya et al., 2000).
Acknowledgement
Encouragements and support from Sachin Purohit and Bikash Aich during the course of the study is gratefully acknowledged.
368
References
- Adamson JW, Fialkow PJ, Murphy S, Prchal JF and Steinmann L (1976). Polycythemia vera: Stem-cell and probable clonal origin of the disease. N Engl J Med 295:913-916.
- Agarwal MB (2007). Clinical Applications of Molecular Haematology: JAK2 in Myeloproliferative Disorders. JAPI 55: 507-510.
- Bartram C, de Klein A, Hagemeijer A. Translocation of c-abl oncogene correlates with the presence of the Philadelphia chromosome in chronic myelocytic leukaemia (1983). Nature 306:277-280.
- Baxter EJ, Scott LM, Campbell PJ, East C, Fourouclas N, Swanton S, Vassiliou GS, Bench AJ, Boyd EM, Curtin N, Scott MA, Erber WN and Green AR (2005). Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet 365:1054-1061.
- Berlin NI (1975). Diagnosis and classification of the polycythemias. Semin Hematol. 12:339-351.
- Correa PN, Eskinazi D and Axelrad AA (1994). Circulating erythroid progenitors in polycythemia vera are hypersensitive to insulin-like growth factor-1 in vitro: Studies in an improved serum-free medium. Blood 83:99-112.
- Dai CH, Krantz SB, Dessypris EN, Means Jr. RT, Horn ST and Gilbert HS (1992). Polycythemia vera. II. Hypersensitivity of bone marrow erythroid, granulocyte-macrophage, and megakaryocyte progenitor cells to interleukin-3 and granulocyte- macrophage colony-stimulating factor. Blood 80:891-899.
- Dai CH, Krantz SB, Green WF and Gilbert HS (1994). Polycythaemia vera. III. Burst-forming units-erythroid (BFU-E) response to stem cell factor and c-kit receptor expression. Br J Haematol 86:12-21.
- Damashek W (1951). Some speculations on the myeloproliferative syndromes. Blood 6:372-375.
- Fialkow PJ, Gartler SM and Yoshida A (1967). Clonal origin of chronic myelocytic leukemia in man. Proc Natl Acad Sci USA 58:1468-1471.
- Gadhia, PK, Vaniawala, S and Pithawala, M (2005). Some Observations on Spontaneous Sister Chromatid Exchange Frequencies and Cell Cycle Progression in Stimulated Lymphocytes of Patients With Different Malignancies. Int J Hum Genet, 5: 187-191.
- James C, Ugo V, Le Couedic JP, Staerk J, Delhommeau F, Lacout C, Garcon L, Raslova H, Berger R, Bennaceur-Griscelli A, Villeval JL, Constantinescu SN, Casadevall N and Vainchenker W (2005). A unique clonal JAK2 mutation leading to constitutive signaling causes polycythaemia vera. Nature 434:1144-1148.
- Jelinek J, Oki Y, Gharibyan V, Bueso-Ramos C, Prchal JT, Verstovsek C, Beran M, Estey E, Kantarjian HM, and Issa JPJ (2005). JAK2 mutation 1849G>T is rare in acute leukemias but can be found in CMML, Philadelphia chromosome negative CML and megakaryocytic leukemia. Blood 05: 1800-1809.
- Khwaja A (2006). The role of Janus kinases in haemopoiesis and haematological malignancy. Br J Haematol. 134:366-384.
- Kralovics R, Passamonti F, Buser AS, Teo SS, Tiedt R, Passweg JR, Tichelli A, Cazzola M and Skoda RC (2005) A gain-of function mutation of JAK2 in myeloproliferative disorders. N Engl J Med 352:1779-1790.
- Lacout C, Pisani DF, Tulliez M, Moreau Gachelin F, Vainchenker W, Villeval JL. JAK2V617F expression in murine hematopoietic cells leads to MPD mimicking human PV with secondary myelofibrosis. Blood 108:1652- 1660.
- Landolfi R (1998). Bleeding and thrombosis in myeloproliferative disorders. Curr Opin Hematol. 5:327-331.
- Levine RL, Wadleigh M, Cools J, Ebert BL, Wernig G, Huntly BJ, Boggon TJ, Wlodarska I, Clark JJ, Moore S, Adelsperger J, Koo S, Lee JC, Gabriel S, Mercher T, D’Andrea A, Frohling S, Dohner K, Marynen P, Vandenberghe P, Mesa RA, Tefferi A, Griffin JD, Eck MJ, Sellers WR, Meyerson M, Golub TR, Lee SJ and Gilliland DG (2005). Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell 7:387-397.
- Ma W, Kantarjian H, Zhang X, Yeh CH, Zhang ZJ, Verstovsek S, Albitar M (2009). Mutation profile of JAK2 transcripts in patients with chronic myeloproliferative neoplasias. J Mol Diagn. 11:49-53
- McWeeneya DO, Galluzzi JR and Ordovas JM (2000). Allelic Discrimination for Single Nucleotide Polymorphisms in the Human Scavenger Receptor Class B Type 1 Gene Locus Using Fluorescent Probes Clinical Chemistry 46: 118-119.
- Mukhopadhyaya PN, Mehta HH and Rathod RN (2000). A method for the simulation of normal, carrier and affected controls for PCR-RFLP screening of genetic disease in dairy cattle. Molecular and Cellular Probes 14:381-384.
- Ohyashiki JH, Hisatomi H, Shimizu S, Sugaya M, Ohyashiki K (2009). Detection of Low Allele Burden of JAK2 Exon 12 Mutations Using TAcloning in Patients with Erythrocytosis. Jpn J Clin Oncol. [Epub ahead of print]
- Parganas E, Wang D, Stravopodis D, Topham DJ, Marine JC, Teglund S, Vanin EF, Bodner S, Colamonici OR, van Deursen JM, Grosveld G and Ihle JN (1998). Jak2 is essential for signaling through a variety of cytokine receptors. Cell 93:385-395.
- Prchal JF and Axelrad AA (1974). Letter: Bone-marrow responses in polycythemia vera. N Engl J Med 290:1382-1389.
- Rapado I, Albizua E, Ayala R, Hernández JA, Garcia-Alonso L, Grande S, Gallardo M, Gilsanz F, Martinez-Lopez J (2008). Validity test study of JAK2 V617F and allele burden quantification in the diagnosis of myeloproliferative diseases. Ann Hematol. 87:741-749.
- Rozen S and Skaletsky H (1999). Primer3 on the WWW for General Users and for Biologist Programmers. Methods in Molecular Biology 132:365- 386.











