Keyword
Cancer, Chemokines, Natural and marine compounds, Maestro 9.6, Molecular docking, In vitro
Introduction
Chemokines are a family of proteins that comprises 44 members in humans, which have a propensity for the migration of cells. Chemokines and their receptors, contributed a significant role in physiological functions such as responses, allowing lymphocyte maturation, cell survival, cell proliferation, and cell-migration, these movement engaged in cancer initiation, angiogenesis, tumor growth, and metastasis. The overexpression of chemokine receptors by blood cancer and solid tumors augment the possibility of cancer initiation and progression [1].
CXCR2 predominant chemokine receptor has been implicated in many cancers including melanoma, breast, prostate, renal, pancreatic, and esophageal cancer. Targeting CXCR2 enhances chemotherapeutic response, inhibits the angiogenesis, and metastasis of breast cancer [2,3]. CXCR4 is the prime chemokine receptor has been alarmed in many cancers including melanoma, breast, ovarian, prostate, pancreatic, colorectal, renal, osteosarcoma, NSCLC, AML, ALL, multiple myeloma [4,5]. Moreover, CXCR2, CXCR4 and CCR5 has a key role in fundamental aspects of cancer, including proliferation, migration, invasion, and angiogenesis [6,7]. Ziconotide, a peptide discovered from marine tropical cone snail, was approved in the United States in December 2004 for the treatment of Prialt (Elan Pharmaceuticals). Moreover, in October 2007, trabectedin (Yondelis; Pharma Mar) became the first marine anticancer drug that was approved in the European Union [8].
Monoclonal antibodies against CXCR2, CXCR4 and CCR5, have entered into clinical trials for cancer therapy including MLN1202 and plerixafor [9,10]. Monoclonal antibodies relatively have better options due to their long half-life in blood, their ability to establish concrete and high-affinity interactions with immune cells, together with their comparatively low toxicity.
But on the other hand, drugs from monoclonal antibodies difficult and expensive to develop, in addition to less convenient to administer than small molecule drugs. Inhibiting the chemokine ligands/receptors interactions present on the tumor cells ensued as a new therapeutic approach. MariaVela 2015 reported that CXCR2, CXCR4, and CCR5 chemokine are most activated chemokine during the metastases in various cancer including melanoma, breast cancer, ovarian cancer, prostate cancer, glioma, neuroblastoma, squamous cell cancer, head and neck cancer [1]. In this context, receptor-based molecular docking was performed to identify multitargeted inhibitor for chemokine receptor.
Methodology
Selection of ligands and protein molecules
In Silico molecular docking protocol adapted from our previous published literature with some modification [11-15] (Figure 1). The database of 200 Marine and natural compounds used in our molecular docking studies were derived from different medicinal plant extracts. Marine and natural compounds selected as ligand molecules that had been reported in the published literature [16-30]. Chemokines receptor molecules selected from published literature [1]. The X-ray three-dimensional crystal structure of chemokines receptor CXCR2 (PDB, 4JL7) [31], CXCR4 (PDB, 3OE6) [32], and CCR5 (PDB, 4MBS) [33] retrieved from the Protein Data Bank (PDB).
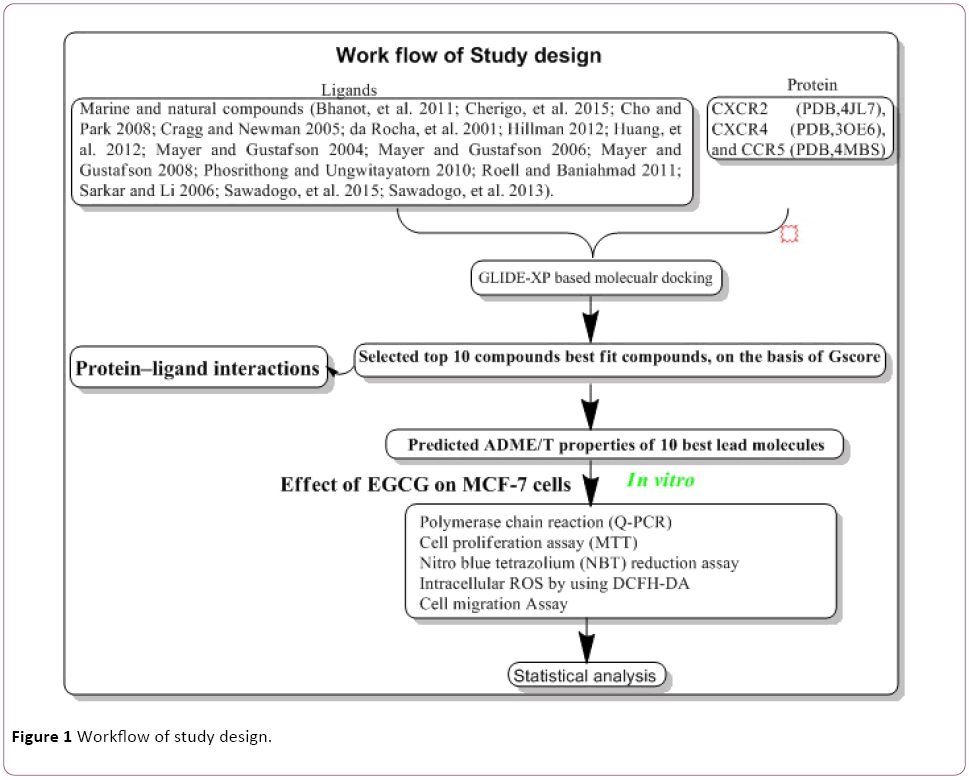
Figure 1: Workflow of study design.
Preparation of ligands molecules
Input ligand molecules were prepared using LigPrep wizard applications where functions such as fixing of charges and orientation of groups, 2D to 3D conversion, corrected bond lengths and bond angles, was incorporated into ligands molecules followed by minimization and optimization was done using OPLS_2005 force field [34-36]. The hydrogen atoms were added to the ligands. Ten tautomers for each ligand generated during ligands preparation.
Preparation of protein molecules
The X-ray three-dimensional crystal structure of selected chemokines receptor CXCR2 (PDB, 4JL7), CXCR4 (PDB, 3OE6), and CCR5 (PDB, 4MBS) were imported to the protein preparation wizard application of maestro 9.6. Preparation and refinement of proteins were done using protein preparation wizard applications where four necessary steps viz. preprocess, review, modify and refinement corrected the bond length, bond angle and others constraint of proteins. Preprocess step made essential modifications such as the addition of hydrogen atoms, assigning bond orders, the creation of disulphide bonds, fixing of the charges and orientation of groups into the raw PDB structure. After that, generate state selection available in the review and the modify tab is executed, considering the pH range of 7+3. Finally, proteins are optimized and minimized with options available in refining panel of protein preparation wizard. Optimization was done with default settings, and then minimization step was executed. All-atom force field (OPSL-2005) applied for charges, and atom types were assigned. After the completion of ligands and protein preparation, a receptor grid file was generated.
GLIDE (grid-based ligand docking with energetics) molecular docking
Molecular docking was performed by employing GLIDE and outputs represented as the Gscore. GLIDE-XP Molecular docking studies using the selected ligand molecules were conducted using Maestro 9.6 molecular docking [37-39]. Each of these selected compounds was docked into target protein molecules and the docking conformation possessing the lowest energy was selected. Molecular docking was performed after receptor-grid generation. The final energy evaluation was done on the basis of Gscore.
Absorption, distribution, metabolism, excretion and toxicity (ADME/T) studies
Drugs don’t accomplish something in clinical trials due to poor compound solubility and gastric emptying time diminishes the extent to which a drug is absorbed after oral administration. Therefore, In Silico ADME/T predictive tools are cooperative strategy that could abolish indecorous compounds, before invested valuable time and money in primary testing of compounds. Thus, ADME properties of bestdocked compounds were predicted using fast mode QikProp application of Maestro 9.6 [40,41].
Reagents
DMEM media, 1% penicillin and streptomycin and fetal bovine serum (FBS) were procured from Invitrogen. EGCG purchased from MP Biomedicals Pvt Ltd and dissolved in dimethylsulfoxide (DMSO) as a 20 MM stock solution. The final dilution was done in culture medium at a where DMSO concentration of 0.25% (vol/vol).
Cell lines
MCF-7 cell line was procured from NCCS, Pune, India. Cells was grown in NCCS recommended phenol red media containing 10% FBS and 1% penicillin and streptomycin have to be maintained in a 37°C incubator with 5% CO2 humidified atmosphere.
Total RNA isolation, cDNA synthesis, and quantitative RT-PCR
One million MCF-7 cells/well was plated in the six-well culture plate and incubated at 37°C overnight with 5% Co2. Cells were treated with EGCG 80 μM for 48 hrs and total RNA was extracted using Tri-reagent (Life Technologies, Gaithersburg, MD). For cDNA synthesis, one μg of total RNA was reverse-transcribed into cDNA using cDNA synthesis kit (Takara Bio Inc). Consequently, mRNA expression was quantified by RT-PCR by using Veriti® 96-well fast thermal cycler (Applied Biosystems). The cycling parameters for PCR were optimized as follows: start at 60°C for 2 minutes, followed by activation at 95°C for 4 minutes, denaturing at 95°C for 20 seconds, annealing at 58°C for 45 seconds, and elongation at 72°C for 45 seconds, repeated for 40 amplification cycles. To confirm correct amplification and size, obtained products of RT-PCR were separated by gel electrophoresis. Further, GAPDH was used as a housekeeping gene to conform equal loading of the sample. Gel band intensities of PCR product was calculated with Bio-Rad's image lab tool.
MTT Assay
Approximately 7 × 103 cells/well cells were seeded in each well in 96-well plates with 10% FBS media after 24 hrs incubation, the media were discarded and added FBS-free media for serum starvation. The cells were exposed to the EGCG containing 5% media and incubated for 48 hrs then media were removed, and 100 μl of MTT (0.5 mg/ml) in PBS was added for 4 hrs. Supernatant was removed and MTT dye was dissolved with 200 μl/well DMSO. Absorbance was measured by using fluorescence microplate reader (Synergy /H1) with a wavelength of 570 nm.
Determination of intracellular reactive oxygen species (ROS)
The levels of ROS were determined quantitatively by using 2, 7-dichloro dihydro fluorescein diacetates (DCFH-DA). 7 × 103 cells/well was plated in the 96-well plate. Media were replaced, in next day with 200 μL of fresh complete medium containing varying concentrations of EGCG after 48 hrs; the supernatants were removed, and re-suspending in 100 μL 5 μM CM-H2DCFDA. Subsequently, the fluorescence intensity was measured at an excitation wavelength of 485 nm and emission wavelength of 530 nm using a fluorescence microplate reader (Synergy /H1).
Nitro blue tetrazolium (NBT) reduction assay
7 x 103 cells/well were seeded into the 96 well tissue culture plates, media containing 10% FBS. After 24 hrs the media were replaced with 200 μL of fresh medium containing 5, 10, 20, 40, and 80 μM concentrations of EGCG. After 48 hrs, the supernatants were removed, and incubated with 100 μL 0.1% NBT. The reduced NBT was solubilized with 100 μL 2 M KOH and 100 μL DMSO for 30 min. The absorbance was measured for each well at 570 nM using an ELISA (Synergy /H1) plate reader.
Cell-migration assay (In vitro wound-healing assay)
Cells were seeded in a six-well plate and grown overnight to the confluence. After 24 hrs, cultures were replaced with fresh medium containing concentration dependent EGCG treatment. Monolayer cells were scratched with a 200 μL pipette tip to create a wound, and kept in an incubator. The rate of wound closure was assessed and photographed after 48 hrs and adding scale bars to images using ImageJ.
Statistical analysis
Statistical values are expressed as the mean ± SEM (SigmaPlot).
Results
Molecular docking of compounds with chemokine receptor
Cancer is heterogeneous disease; with various cross signaling pathways involved in disease progression. When blocking the rational targeted pathways, the single targeted compounds can also be actuated by interrelated signaling pathways that led to mounting of cell proliferation. Therefore, the multitargeted drugs have the better mechanism of action against the anticancer activity. Multi-target therapeutics involves discovering a single compound that can be active on two or more targets. It is accepted that this approach is biologically more important and can be designed because ADMET properties of a single agent targeted to various protein are easy to predict than the use of two or more singletargeted agents [12,42]. Furthermore, various small molecular chemokines receptor inhibitor such as CXCR4 (burixafor and BKT140) [43,44], CCL2 – CCR2 (cenicriviroc, PF 4136309, and MK 0812) [45], and CCR5 (Maraviroc) have discovered by using In Silico and In vitro technique, and entered in to cancer clinical trials [46]. Moreover, anibamine is the first natural product reported as a CCR5 antagonist and has been identified for anticancer activity [47,48]. In the current study, we performed multitargeted molecular docking of chemokine receptor CXCR2 (PDB, 4JL7), CXCR4 (PDB, 3OE6), and CCR5 (PDB, 4MBS) against natural compounds. The lowest energy conformation, on behalf of the best binding structure of inhibitors to receptor was choosen. The protein-ligands interactions yielded a cornucopia of information highlighting the irrefutable role portrayed by various factors such as hydrogen bonds, salt bridges, metal interactions, lipophilic interactions, π-π and π- cation interactions in the protein-ligand interactions profile. The data achieved from molecular docking various parameter including their Gscore, hydrogen bond, electrostatic bond, and amino acid residues are illustrated in Tables 1 and 2. In the present study, CID56835050, 6918506, 441794, 42607750, 42607750-2, 65064, 72277, 6711419, 42607750-3, and 5372499 showing high Gscore -7.62. -7.2, -7.09, -6.93, -6.84, -6.62, -6.59, -6.23, -6.22, and -6.13 kilocalorie per mole for CXCR2. Hydrophobic interactions are crucial that are involved in protein-ligands interactions. Interestingly, CID56835050 showed the highest binding energy (XP- Gscore) score -7.62 kcal/mol when docked with CXCR2. To see their interacting pattern, docking pose analysis revealed that amino acids Asn22, Gly25, Phe26, and Leu28 formed four backbone hydrogen bonds interactions at the ATP binding site of CXCR2. Moreover, CID65064 showed Gscore -6.62 kcal/mol when docked with CXCR2 and amino acids Tyr24, Phe26, and Hid27 involved in backbone hydrogen bonds in addition to pi-pi interactions (Asn22, Gly25, and Leu28) (Figures 2 and 3).
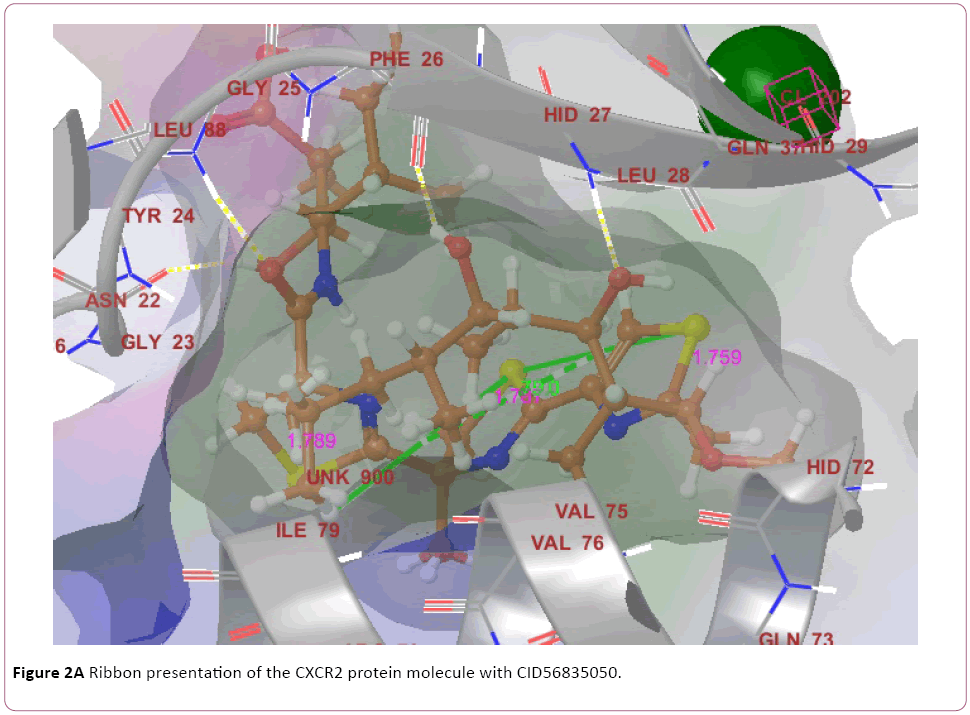
Figure 2a: Ribbon presentation of the CXCR2 protein molecule with CID56835050.
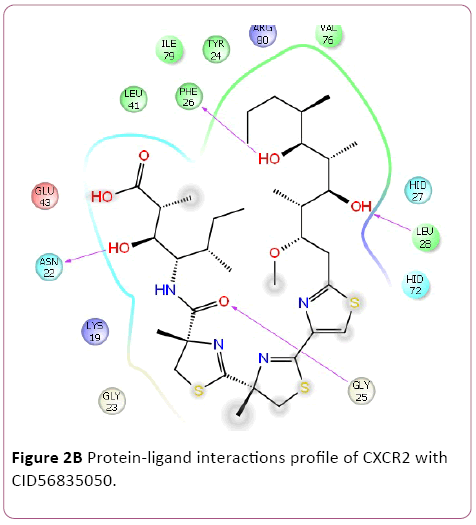
Figure 2b: Protein-ligand interactions profile of CXCR2 with CID56835050.
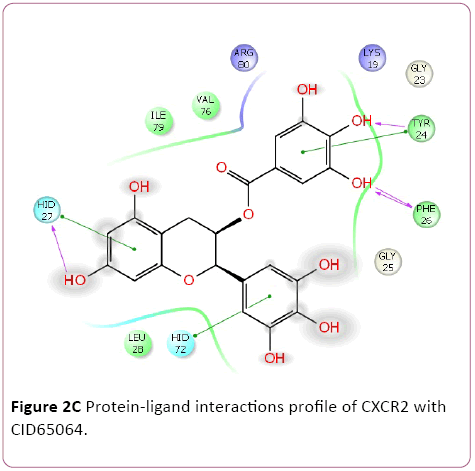
Figure 2c: Protein-ligand interactions profile of CXCR2 with CID65064.
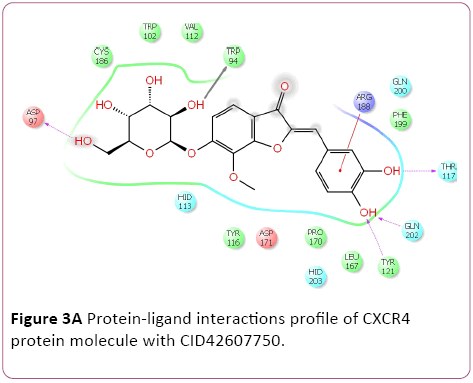
Figure 3a: Protein-ligand interactions profile of CXCR4 protein molecule with CID42607750.
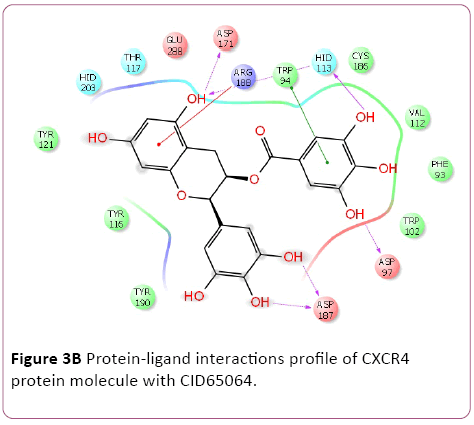
Figure 3b: Protein-ligand interactions profile of CXCR4 protein molecule with CID65064.
| S.N. |
Ligands CID |
GScore |
Lipophilic EvdW |
HBond |
Electro |
Protein-Ligands interactions |
| CXCR2 |
9841667 |
-4.9 |
-2.5 |
-1.76 |
-0.72 |
Tyr24, Phe26, Hid27, and Arg80 |
| |
3854666 |
-4.73 |
-2.37 |
-1.28 |
-0.73 |
Asn22, Gly25, and Glu43 |
| |
10474776 |
-4.3 |
-2.86 |
-0.92 |
-0.44 |
Tyr24, Gly25, and Arg80 |
| |
11256587 |
-3.8 |
-2.8 |
-0.28 |
-0.15 |
Phe87 and Asp119 |
| |
11565518 |
-3.39 |
-2.3 |
-0.85 |
-0.49 |
Phe87, Trp94, and Tyr116, |
| CXCR4 |
483559 |
-2.84 |
-2.7 |
-0.38 |
-0.26 |
Ser123 |
| |
65015 |
-2.45 |
-2.02 |
-0.35 |
-0.23 |
Asp84 and Phe87 |
| |
3002977 |
-8.79 |
-5.22 |
-0.45 |
-0.61 |
Tyr37, Phe109, Phe112, and Tyr251 |
| |
3073217 |
-6.38 |
-4.1 |
-1.75 |
-0.54 |
Ser180 |
| CCR5 |
3001322 |
-6.24 |
-3.61 |
-0.96 |
-0.48 |
Lys22, Lys191, and Glu162 |
| |
3009355 |
-5.91 |
-3.24 |
0 |
-0.08 |
Tyr108 |
| |
54682467 |
-5.09 |
-2.06 |
-0.65 |
-0.22 |
Tyr251 |
Table 1: Lowest binding energy for the ligand (controls)-CXCR2, CXCR4, and CCR5 protein interactions.
| Proteins |
Com. CID |
Gscore |
Lipophilic E vdw |
HBond |
Electro |
Protein-ligands interactions |
| CXCR2 |
56835050 |
-7.62 |
-2.87 |
-3.92 |
-1.07 |
Asn22, Gly25, Phe26, and Leu28 |
| 6918506 |
-7.2 |
-1.97 |
-2.58 |
-0.89 |
Gly25, Phe26, Leu28, and Arg80 |
| 441794 |
-7.09 |
-1.72 |
-4.67 |
-0.84 |
Asn22, Gly25, Phe26, Hid27, and Leu28 |
| 42607750 |
-6.93 |
-1.26 |
-4.27 |
-1.66 |
Asn22, Gly25, Phe26, Hid27, Leu28, and Arg80 |
| 42607750-2 |
-6.84 |
-1.27 |
-4.1 |
-1.7 |
Asn22, Tyr24, Hid27, Leu28, Hid72, and Arg80 |
| 65064 |
-6.62 |
-2.03 |
-3.59 |
-0.91 |
Tyr24, Hid27, and Hid72, |
| 72277 |
-6.59 |
-1.32 |
-3.92 |
-1.22 |
Asn22, Gly25, Phe26, and Arg80 |
| 6711419 |
-6.23 |
-3.13 |
-2.22 |
-1.17 |
Asn22, Gly25, Phe26, and Arg80 |
| 42607750-3 |
-6.22 |
-2.03 |
-3.44 |
-0.98 |
Tyr24, Leu28, and Glu43 |
| 5372499 |
-6.13 |
-1.38 |
-1.84 |
-0.53 |
Asn22, Gly25, and Phe26, |
| CXCR4 |
42607750 |
-10.87 |
-4.16 |
-4.3 |
-1.34 |
Trp94, Asp97, Thr117, Tyr121, Arg188, and Gln202 |
| 42607750-2 |
-9.53 |
-3.07 |
-4.38 |
-1.75 |
Trp94, Asp97, Tyr116, Phe199, Gln202, and Tyr 225 |
| 6441009 |
-9.46 |
-3.87 |
-4.07 |
-1.44 |
Asp97, Tyr116, Phe 199, Gln202, Tyr 225, Glu288 |
| 42607750-3 |
-9.28 |
-3.43 |
-4.5 |
-1.26 |
Hid113, Thr117, Tyr121, and Cys186 |
| 42607750-4 |
-9.21 |
-3.46 |
-4.18 |
-1.73 |
Asp97, Tyr116, Phe199, Gln202, and Arg188 |
| 42607750-5 |
-8.8 |
-3.01 |
-3.84 |
-1.76 |
Asp97, Tyr116, Phe199, and Gln200 |
| 42607750-6 |
-8.66 |
-3.54 |
-4.18 |
-1.05 |
Asp97, Tyr116, Phe199, Gln202, and Arg188 |
| 16075395 |
-8.56 |
-4.81 |
-3.16 |
-0.89 |
Thr117, Tyr121, Hid113, and Hid203 |
| 65064 |
-8.47 |
-3.31 |
-3.67 |
-1.78 |
Trp94, Asp97, Hid113, Asp171, Asp187, and Arg188 |
| 70690672 |
-8.29 |
-3.64 |
-3.52 |
-1.06 |
|
| CCR5 |
6441009 |
-10.74 |
-3.24 |
-5.05 |
-1.25 |
Ser180, Tyr187, Asp276, and Glu283 |
| 636710 |
-10.62 |
-6.58 |
-1.13 |
-0.61 |
Tyr89, Phe109, and Ser180 |
| 6441009-2 |
-10.28 |
-4.13 |
-4.4 |
-0.44 |
Tyr87 and Thr195 |
| 42607750 |
-9.62 |
-3.35 |
-4.49 |
-0.93 |
Tyr37, Trp86, Tyr108, and Gln280 |
| 6473739 |
-9.6 |
-3.35 |
-3.53 |
-1.17 |
Ser180, Glu283 and Hid181 |
| 6441009-3 |
-9.49 |
-2.86 |
-4.15 |
-1.17 |
Ser180, Glu283 and Hid181 |
| 6441009-4 |
-9.42 |
-2.34 |
-4.57 |
-1.62 |
Ser180, Asp276, Glu283 and Hid181 |
| 65064 |
-9.28 |
-2.58 |
-4.71 |
-1.59 |
Tyr37, Tyr108, and Glu283 |
| 6441009-5 |
-9.28 |
-2.99 |
-3.84 |
-1.29 |
Asp276, and Glu283 |
| 56835050 |
-9.21 |
-5.67 |
-2.38 |
-0.91 |
Phe109 |
Table 2: Lowest binding energy for the ligand-CXCR2, CXCR4, and CCR5 protein interactions.
Molecular docking result of CXCR4 against natural compounds revealed that CID42607750, 42607750-2, 6441009, 42607750-3, 42607750-4, 42607750-5, 42607750-6, 16075395, 65064, and 70690672 have better Gscore, -10.87, -9.53, -9.46, -9.28, -9.21, -8.8, -8.66, -8.56, -8.47, and -8.29 Kcal/mol respectively. Protein-ligands interactions pattern showed that amino acid Trp94, Asp97, Thr117, Tyr121, Arg188, and Gln202 involved in different type of bond formation such as hydrogen bond, pi-cation interactions and pi-pi interactions. Furthermore, it has showed that amino acid Trp94, Asp97, Hid113, Asp171, and Asp187 involve inside chain hydrogen bond formation whereas Hid113 formed backbone hydrogen bond when docked with CID65064. Moreover, Arg188 intricated in noncovalent molecular interaction such as cation– π interactions in addition to pi-pi interactions (Trp94) with CID65064. Furthermore, compounds CID6441009, 636710, 6441009-2, 42607750, and 6473739 showed high binding affinity Gscore -10.74, -10.62, -10.28, -9.62, -9.6 respectively when docked with CCR5. Amino acids Tyr37, Trp86, Tyr89, Phe109, Ser180, Tyr187, Asp276, and Glu283 occupied in different bond formation during interactions (Figure 4).
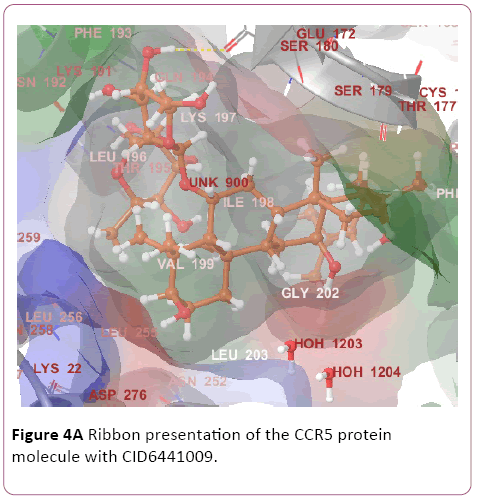
Figure 4a: Ribbon presentation of the CCR5 protein molecule with CID6441009.
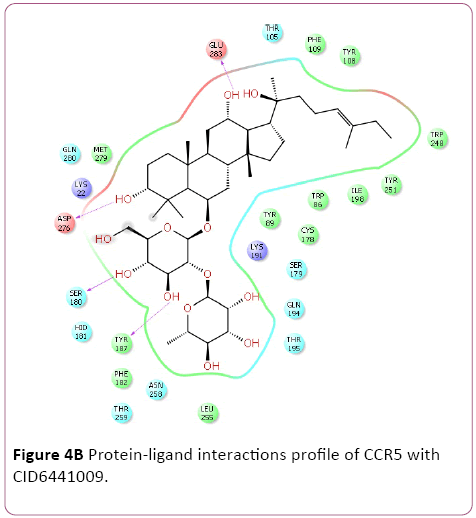
Figure 4b: Protein-ligand interactions profile of CCR5 with CID6441009.
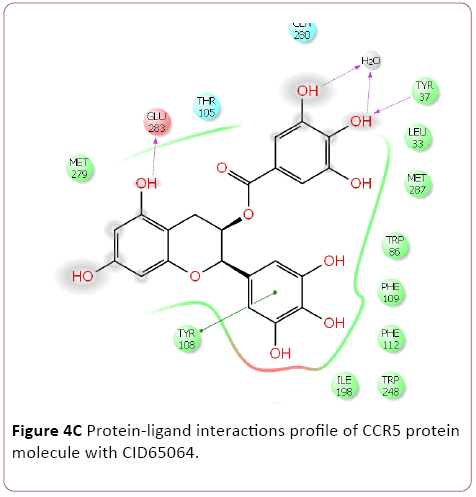
Figure 4c: Protein-ligand interactions profile of CCR5 protein molecule with CID65064.
ADME/T Studies
The ligands were evaluated for drug-likeness of the lead molecules by appraises their physicochemical properties through the Qikprop application of Maestro 9.6. These are fundamental physicochemical properties that are prerequisite for drug development such as compounds have molecular weights were <500 daltons, <5 hydrogen bond donors, <10 hydrogen-bond acceptors and a logp of <5. Our lead molecules compounds have the acceptable range of the physicochemical properties of drug-like molecules. Moreover, these compounds were further evaluated for pharmacokinetic parameters required for absorption, distribution, metabolism, and excretion (ADME). All lead compounds, the partition coefficient (QPlogPo/w) and QPlog Herg, critical for estimation of absorption and distribution of drugs within the body, ranged between −2 to 6.5 and, above -5.0 respectively. Further, QPP Caco, QP log BB, QPP MDCK, QPlog KP, and QPlog Khsa all these pharmacokinetic parameters were within the acceptable range defined for human use, thereby indicating their potential as drug-like molecules (Table 3).
Compounds
CID |
QP log
P o/w
(-2.0
to 6.5) |
QPlog HERG
(acceptable range: above -5.0) |
QPP Caco
(nm/sec)
<25-poor
>500- great |
QP log
BB
(-3-1.2) |
QPP
MDCK
(nm/sec)
<25-poor
>500- great |
QPlog
Kp
(-8.0
to -0.1) |
QPlogKhsa
(Acceptable range: -1.5 to 1.5). |
Percentage of human oral absorption; (<25% is poor and >80% is high) |
| 56835050 |
4.85 |
-1.49 |
27.18 |
-2.31 |
62.05 |
-2.97 |
-0.01 |
55.13 |
| 6918506 |
1.65 |
-4.03 |
814.90 |
-1.22 |
396.52 |
-2.46 |
-0.46 |
62.82 |
| 441794 |
0.84 |
-3.41 |
96.92 |
-1.69 |
39.70 |
-4.56 |
-0.27 |
67.45 |
| 42607750 |
-0.55 |
-5.66 |
13.18 |
-3.26 |
4.59 |
-5.20 |
-0.88 |
17.81 |
| 42607750 |
-0.58 |
-5.67 |
12.27 |
-3.30 |
4.25 |
-5.27 |
-0.89 |
17.07 |
| 65064 |
-0.51 |
-5.15 |
0.75 |
-4.19 |
0.21 |
-7.86 |
-0.46 |
0 |
| 72277 |
-0.33 |
-4.13 |
19.24 |
-2.24 |
6.91 |
-5.69 |
-0.57 |
35.018 |
| 6711419 |
1.16 |
-2.98 |
1126.43 |
-0.90 |
815.86 |
-1.93 |
-0.94 |
62.44 |
| 42607750 |
-0.58 |
-5.50 |
15 |
-3.13 |
5.28 |
-5.11 |
-0.89 |
18.68 |
| 5372499 |
-0.23 |
-3.43 |
358.45 |
-0.87 |
163.20 |
-3.84 |
-0.75 |
71.29 |
| 56835050 |
4.85 |
-1.49 |
27.18 |
-2.31 |
62.05 |
-2.97 |
-0.01 |
55.13 |
| 6918506 |
1.65 |
-4.03 |
814.90 |
-1.22 |
396.52 |
-2.46 |
-0.46 |
62.82 |
| 6441009 |
1.47 |
-5.33 |
12.35 |
-4.04 |
4.28 |
-5.30 |
-0.43 |
16.27 |
| 636710 |
3.76 |
-2.97 |
161.97 |
-1.53 |
138.21 |
-2.66 |
-0.53 |
62.60 |
| 6473739 |
6.37 |
-6.24 |
17.80 |
-4.69 |
6.35 |
-4.19 |
1.05 |
47.79 |
| 6400741 |
3.50 |
-6.93 |
13.75 |
-3.87 |
44.70 |
-4.43 |
-0.0 |
41.93 |
Table 3: Evaluation of drug-like properties of the lead molecules by Qikprop Maestro.
Effect of EGCG on mRNA expression of chemokine receptor in MCF-7 cells
We investigated whether EGCG could hamper chemokine receptor expression in MCF-7 cells. Interestingly, RTPCR shows that EGCG have a potential for inhibition of CXCR2 and CCR5 in 40 μM and 80 μM EGCG. Furthermore, pro-apoptotic proteins Bid mRNA expression was determined by RTPCR. EGCG induce the pro-apoptotic proteins (Bid) expression (Figure 5).
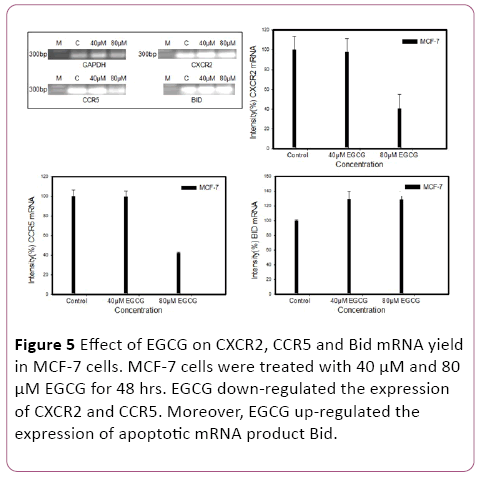
Figure 5: Effect of EGCG on CXCR2, CCR5 and Bid mRNA yield in MCF-7 cells. MCF-7 cells were treated with 40 μM and 80 μM EGCG for 48 hrs. EGCG down-regulated the expression of CXCR2 and CCR5. Moreover, EGCG up-regulated the expression of apoptotic mRNA product Bid.
Effects of EGCG on cell growth of MCF-7 cells
To measure the cytotoxicity of EGCG in MCF-7 cells, MTT assay was performed. Cells were cultured with 5, 10, 20, 40, and 80 μM EGCG for 48 hrs. EGCG exhibited a remarkable reduction of cell proliferation against MCF-7 cells in a concentration-dependent manner. Consistently, it was more prominent at concentrations between 20 μM to 80 μM. The mean 50% inhibitory concentration of EGCG on cell growth was at 20 μM on MCF-7 cells for 48 hrs treatment (Figure 6).
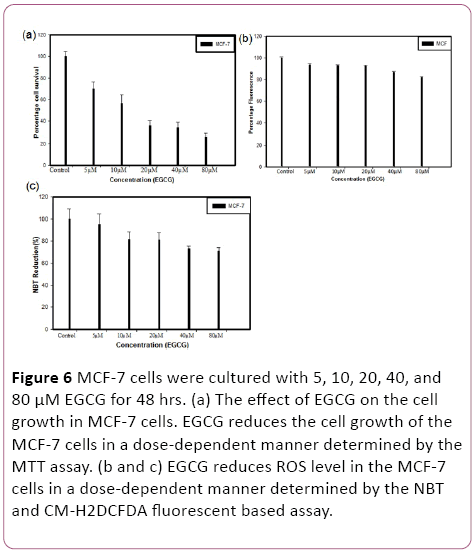
Figure 6: MCF-7 cells were cultured with 5, 10, 20, 40, and 80 μM EGCG for 48 hrs. (a) The effect of EGCG on the cell growth in MCF-7 cells. EGCG reduces the cell growth of the MCF-7 cells in a dose-dependent manner determined by the MTT assay. (b and c) EGCG reduces ROS level in the MCF-7 cells in a dose-dependent manner determined by the NBT and CM-H2DCFDA fluorescent based assay.
The effects of EGCG on reactive oxygen species (ROS) by fluorescent DCF
EGCG exposure led to significant changes in relative ROS were observed on MCF-7 cell line after 48 hrs treatments. MCF-7 cells were cultured with 5, 10, 20, 40, and 80 μM EGCG for 48 hrs and calculated the ROS reduction. The mean 20% ROS reduction at 80 μM on MCF-7 cells.
The effects of EGCG on Nitroblue tetrazolium (NBT) reduction on MCF-7 cells
We assessed the ability of the EGCG to initiate the generation of ROS in MCF-7 cells by means of the colorimetric nitroblue-tetrazolium (NBT) assay. In MCF-7 cells, the EGCG caused a reduction in ROS generation that led to reduced cell proliferation.
EGCG possesses migration-suppressing potential
A wound-healing assay was used to evaluate the effect of EGCG on metastatic activity (cell-migration). The treatment of MCF-7 cells with 5 μM to 80 μM EGCG for 48 hrs resulted in a reduction in the degree of wound healing, indicating that EGCG are able to inhibit metastatic activity in MCF-7 cells (Figure 7).
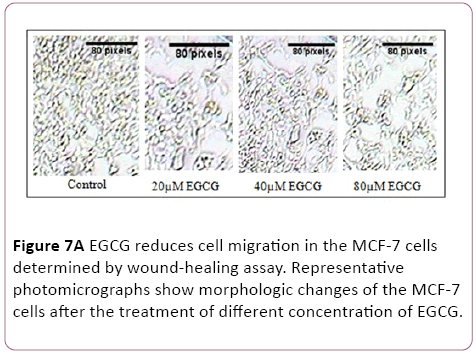
Figure 7a: EGCG reduces cell migration in the MCF-7 cells determined by wound-healing assay. Representative photomicrographs show morphologic changes of the MCF-7 cells after the treatment of different concentration of EGCG.

Figure 7b: The rate of wound closure was photographed after 48 hrs and adding scale bars to images using ImageJ.
Discussion
Chemokines are categorized in four major class on the basis of number and spacing of conserved cysteine residues on their N-terminus viz; CXC, CC, CX3C, and XC. A family of 7- transmembrane spanning G-protein-coupled receptors mediates the attractant effects of chemokines. Expressions of chemokines receptors portray a dual role in tumor initiation and progression. 1) Anti-tumor immunity, chemokines secreted by normal or cancerous cells can help to increase leukocyte migration toward the site of the requirement that led to anticancer activity. 2) They may facilitate survival, proliferation, and metastatic potential of tumor cells. Many chemokines are proinflammatory for most tumors and have essential functions in tumor angiogenesis. In recent years, natural compounds have been investigated tremendously in the anticancer properties. Some promising new agents are in clinical development based on discriminatory activity against anticancer molecular targets. Natural compounds such as quercetin and genistein have entered late phase clinical trials accredited to their oncological indications with the absence of recognizable side effects. At the molecular level, natural compounds have been reported to modulate several protein kinases including RTK, G-protein coupled receptor, and lipoxygenase that plays a significant role in cancer pathology.
Last few years it is assumed that inhibiting the p53-MDM2 interactions are one of the most promising targets for cancer therapy. Hoiamide D (CID56835050) a marine cyanobacteriaderived compound which are involved in inhibition of p53/ MDM2 interaction and have best binding energy with chemokines receptor, it may be an attractive anticancer compounds [49]. Moreover, peloruside A (CID6918506) is a secondary metabolite isolated from New Zealand marine sponges that have the antimitotic agent. Peloruside A has effective microtubule-stabilizing activity and is cytotoxic at nanomolar concentrations. Peloruside A arrests cells in the G2- M phase of the cell cycle and induces apoptosis [50]. Moreover, peloruside A was shown to induce the formation of micronuclei, and microtubule bundles fibers that induce cellcycle arrest in the G2-M phase and apoptosis [51]. Glaucarubin (CID441794) isolated from the tropical plant, simarouba glauca and used to treat cancer growth. Among various docked compound EGCG seems to be a good lead molecule which represents the binding energy of -6.62 Kcal/mol in the case of CXCR2 that is good compared to standard compounds. Furthermore, EGCG also has good binding ability towards CXCR4, having the binding energy of -8.47 Kcal/mol which is comparatively better than the binding energy of standard compounds. Furthermore, in the case of CXCR5 the binding energy of EGCG was found to be -9.28 Kcal/mol which is comparable better than standard compounds. Thus, compound EGCG appears to be a good lead molecule which modulates the activity of CXCR2, CXCR4 as well as CCR5. EGCG down-regulates inflammatory cytokine in HT29 and T84 colon cancer cells [52]. Moreover, it was shown that antimetastatic effects of EGCG on uveal melanoma cells via the downregulation of MMP2 [53]. Furthermore, EGCG treatment in periodontitis inhibits CCL11 production [54]. It has shown that anti-inflammatory EGCG bound directly to chemokines CXCL9, CXCL10, and CXCL11 and limiting their inflammatory activity [55]. Myricetin showed protective effect of diet-induced obesity and insulin resistance by regulating inflammatory responses via blocking AKT and p38 activation [56]. Ginsenoside, (CID6441009) one of the energetic ingredient of Panax ginseng, has a multiple physiologic and pharmacologic effects. Ginsenoside Rd inhibits the proliferation and survival of gastric and breast cancer cells by inhibiting melastatin type transient receptor potential 7 (TRPM7) channel activity [57]. Furthermore, it has shown that ginsenoside, administration with other anti-cancer agents may be useful for the treatment of chemotherapy-resistant breast cancer through downregulating MDR1 [58]. Moreover, Ginsenoside Rd in HT29 was involved in cytoprotection, DNA replication and repair, protein synthesis and degradation, metastasis and mutagenesis that supposed that ginsenoside Rd contributed to induce anticancer activity [59]. In vitro and in vivo studies have established an irrefutable association between natural products induced modulation of protein kinase and MMPs activities with apoptosis, cellular proliferation, tumor cell invasive and inhibition of angiogenesis. Therefore, we can conclude that promotion of natural products for a preferred anticancer strategy to decrease the burden of cancer.
Conclusion
Natural and marine compounds modulate many biological actions associated with cancer progression and development including the cell proliferation, apoptosis, cell differentiation and neovascularization. Hoiamide D, peloruside A, glaucarubin, EGCG, and ginsenoside Rd, have the best docking free energy score against chemokines receptor and some biological descriptors with relevance to (Lipinski rule of five) of the compounds were found to be satisfactory for developing into therapeutics. Protein-ligand interactions profile highlighted that lipophilic interactions, hydrogen bonding, π- cation, and π-π stacking interactions contributed to the active site of proteins that are essential to augment the anticancer activity. Moreover, RT-PCR densitometric bands analysis showed that compound EGCG reduced the mRNA expression of CXCR2, CCR5 and increased the Bid at 40 μM and 80 μM concentration. Moreover, EGCG significantly reduced cell proliferation, ROS generation, and cell-migration after 48 hrs treatments. Further, experimental studies are required for the experimental validation of our findings.
Acknowledgment
We would like to thank Vice Chancellor, Central University of Punjab, Bathinda, Punjab, (India) for supporting this study with infrastructural requirements.
17564
References
- Vela M, Aris M, Llorente M, Garcia-Sanz J, Kremer L (2015) Chemokine receptor specific antibodies in cancer immunotherapy: achievements and challenges. Front Immunol 6:12
- Sharma B, Nawandar DM, Nannuru KC, Varney ML, Singh RK (2013) Targeting CXCR2 enhances chemotherapeutic response, inhibits mammary tumor growth, angiogenesis, and lung metastasis. Mol Cancer ther 12: 799-808
- Sharma B, Singh S, Varney ML, Singh RK (2010) Targeting CXCR1/CXCR2 receptor antagonism in malignant melanoma. Expert OpinTher Targets 14: 435-442
- Müller A, Homey B, Soto H, Ge N, Catron D, et al. (2001) Involvement of chemokine receptors in breast cancer metastasis. Nature 410:50-56.
- Kang H, Watkins G, Douglas-Jones A, Mansel RE, Jiang WG (2005) The elevated level of CXCR4 is correlated with nodal metastasis of human breast cancer. The Breast 14: 360-367.
- Balkwill F (2004) Cancer and the chemokine network. Nat Rev Cancer 4:540-550.
- Ward S, Li K, Hepburn E, Weston C, Curbishley S, et al. (2015) The effects of CCR5 inhibition on regulatory T-cell recruitment to colorectal cancer. Br J Cancer 112: 319-328.
- Molinski TF, Dalisay DS, Lievens SL, Saludes JP (2009) Drug development from marine natural products. Nat Rev Drug Discov 8: 69-85.
- Lu X, Kang Y (2009) Chemokine (CC motif) ligand 2 engages CCR2+ stromal cells of monocytic origin to promote breast cancer metastasis to lung and bone. J Biol Chem 284: 29087-29096.
- Ma Y, Adjemian S, Galluzzi L, Zitvogel L, Kroemer G (2014) Chemokines and chemokine receptors required for optimal responses to anticancer chemotherapy. Oncoimmunology 3.
- Singh P, Bast F (2015) High-throughput virtual screening, identification and In vitro biological evaluation of novel inhibitors of signal transducer and activator of transcription 3. Med Chem Res 1-15.
- Singh P, Bast F (2014) Multitargeted molecular docking study of plant-derived natural products on phosphoinositide-3 kinase pathway components. Med Chem Res 23:1690-1700.
- Singh P, Bast F (2014) In Silico molecular docking study of natural compounds on wild and mutated epidermal growth factor receptor. Med Chem Res1-12.
- Singh P, Bast F (2015) Screening and biological evaluation of myricetin as a multiple target inhibitor insulin, epidermal growth factor, and androgen receptor; In Silico and In vitro. Invest New Drugs 33:575-593.
- Singh P, Bast F (2015) Screening of multi-targeted natural compounds for receptor tyrosine kinases inhibitors and biological evaluation on cancer cell lines, In Silico and In vitro. Med Oncol 32:1-18.
- da Rocha AB, Lopes RM, Schwartsmann G (2001) Natural products in anticancer therapy. Curr Opinion Pharmacol 1:364-369.
- Sarkar FH, Li Y (2006) Using chemopreventive agents to enhance the efficacy of cancer therapy. Cancer Res 66:3347-3350.
- Hillman GG (2012) Dietary Agents in Cancer Chemoprevention and Treatment. J Oncol 21: 209-219.
- Roell D, Baniahmad A (2011) The natural compounds atraric acid and N-butylbenzene-sulfonamide as antagonists of the human androgen receptor and inhibitors of prostate cancer cell growth. Mol Cell Endocrinol 332:1-8.
- Huang W, He T, Chai C, Yang Y, Zheng Y, et al. (2012)Triptolide inhibits the proliferation of prostate cancer cells and down-regulates SUMO-specific protease 1 expression. Pub Libr Sci One 7:e37693.
- Phosrithong N, Ungwitayatorn J (2010) Molecular docking study on anticancer activity of plant-derived natural products. Med Chem Res 19:817-835.
- Cho JY, Park J (2008) Contribution of natural inhibitors to the understanding of the PI3K/PDK1/PKB pathway in the insulin-mediated intracellular signaling cascade. Int J Mol Sci 9:2217-2230.
- Bhanot A, Sharma R, Noolvi MN (2011) Natural sources as potential anti-cancer agents: A review. Int J Mol Phytomed 3:9-26.
- Cragg GM, Newman DJ (2005) Plants as a source of anti-cancer agents. J Ethnopharmacol 100:72-79.
- Mayer AM, Gustafson KR (2008) Marine pharmacology in 2005–2006: Antitumour and cytotoxic compounds. Eur J Cancer 44:2357-2387.
- Mayer AM, Gustafson KR (2006) Marine pharmacology in 2003–2004: anti-tumourand cytotoxic compounds. Eur J Cancer 42:2241-2270.
- Mayer AM, Gustafson KR (2004) Marine pharmacology in 2001–2: antitumourand cytotoxic compounds. Eur J Cancer 40:2676-2704.
- Cherigo L, Lopez D, Martinez-Luis S (2015) Marine Natural Products as Breast Cancer Resistance Protein Inhibitors. Marine Drugs 13:2010-2029.
- Sawadogo WR, Boly R, Cerella C, Teiten MH, Dicato M, Diederich M (2015) A Survey of Marine Natural Compounds and Their Derivatives with Anti-Cancer Activity Reported in 2012. Molecules 20:7097-7142
- Sawadogo WR, Schumacher M, Teiten M-H, Cerella C, Dicato M, et al. (2013) A survey of marine natural compounds and their derivatives with anti-cancer activity reported in 2011. Molecules 18:3641-3673.
- Lu G, Wu Y, Jiang Y, Wang S, Hou Y, et al (2013) Structural insights into neutrophilic migration revealed by the crystal structure of the chemokine receptor CXCR2 in complex with the first PDZ domain of NHERF1. PloS One 8:e76219.
- Wu B, Chien EY, Mol CD, Fenalti G, Liu W, et al (2010) Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science 330:1066-1071.
- Tan Q, Zhu Y, Li J, Chen Z, Han GW, et al (2013) Structure of the CCR5 chemokine receptor–HIV entry inhibitor maraviroc complex. Science 341:1387-1390.
- Shivakumar D, Williams J, Wu Y, Damm W, Shelley J, et al. (2010) Prediction of absolute solvation free energies using molecular dynamics free energy perturbation and the OPLS force field. J Chem Theor Comput 6:1509-1519.
- Jorgensen WL, Maxwell DS, Tirado-Rives J (1996) Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J Am Chem Soc 118:11225-11236.
- Jorgensen WL, Tirado-Rives J (1988) The OPLS [optimized potentials for liquid simulations] potential functions for proteins, energy minimizations for crystals of cyclic peptides and crambin. J Am Chem Soc 110:1657-1666.
- Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, et al (2006) Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J Med Chem 49:6177-6196.
- Halgren TA, Murphy RB, Friesner RA, Beard HS, Frye LL, et al (2004) Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J Med Chem 47:1750-1759.
- Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, et al (2004) Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem 47:1739-1749.
- Lu JJ, Crimin K, Goodwin JT, Crivori P, Orrenius C, et al (2004) Influence of molecular flexibility and polar surface area metrics on oral bioavailability in the rat. J Med Chem 47:6104-6107.
- Jorgensen WL, Duffy EM (2002) Prediction of drug solubility from structure. Ad Drug Del Rev 54:355-366.
- Zhou S, Li Y, Hou T(2013) Feasibility of using molecular docking-based virtual screening for searching dual target kinase inhibitors. J Chem Inform Mod 982-996.
- Peled A, Abraham M, Avivi I, Rowe JM, Beider K, et al (2014) The high-affinity CXCR4 antagonist BKT140 is safe and induces a robust mobilization of human CD34+ cells in patients with multiple myeloma. Clin Cancer Res 20:469-479.
- Debnath B, Xu S, Grande F, Garofalo A, Neamati N (2013) Small molecule inhibitors of CXCR4. Theranostics 3:47-75.
- Sanford DE, Belt BA, Panni RZ, Mayer A, Deshpande AD, et al (2013) Inflammatory monocyte mobilization decreases patient survival in pancreatic cancer: a role for targeting the CCL2/CCR2 axis. Clin Cancer Res 19:3404-3415.
- Adams JL, Smothers J, Srinivasan R, Hoos A (2015) Big opportunities for small molecules in immuno-oncology. Nat Rev Drug Discov 14:603-622.
- Zhang Y, Arnatt CK, Zhang F, Wang J, Haney KM, et al. (2012) The potential role of anibamine, a natural product CCR5 antagonist, and its analogues as leads toward development of anti-ovarian cancer agents. Bioorg Med Chem Lett 22:5093-5097.
- Zhang F, Arnatt CK, Haney KM, Fang HC, Bajacan JE, et al (2012) Structure activity relationship studies of natural product chemokine receptor CCR5 antagonist anibamine toward the development of novel anti prostate cancer agents. Eur J Med Chem 55:395-408.
- Malloy KL, Choi H, Fiorilla C, Valeriote FA, Matainaho T, et al. (2012) Hoiamide D, a marine cyanobacteria-derived inhibitor of p53/MDM2 interaction. Bioorg Med Cchem Lett 22:683-688.
- Hood KA, West LM, Rouwé B, Northcote PT, Berridge MV, et al. (2002) Peloruside A, a novel antimitotic agent with paclitaxel-like microtubule-stabilizing activity. Cancer Res 62:3356-3360.
- Jin M, Taylor RE (2005) Total synthesis of (+)-peloruside A. Organic letters 7:1303-1305.
- Porath D, Riegger C, Drewe J, Schwager J (2005) Epigallocatechin-3-gallate impairs chemokine production in human colon epithelial cell lines. J Pharmacol Exp Ther 315:1172-1180.
- Chang CW, Hsieh YH, Yang WE, Yang SF, Chen Y, et al. (2014) Epigallocatechingallate Inhibits Migration of Human Uveal Melanoma Cells via Downregulation of Matrix Metalloproteinase-2 Activity and ERK1/2 Pathway. BioMed Res Int.
- Hosokawa Y, Hosokawa I, Shindo S, Ozaki K, Matsuo T (2012) (-)-Epigallocatechin-3-gallate inhibits CC chemokine ligand 11 production in human gingival fibroblasts. Cell PhysiolBiochem 31:960-967.
- Qin S, Alcorn JF, Craigo JK, Tjoeng C, Tarwater PM, et al (2011) Epigallocatechin-3-gallate reduces airway inflammation in mice through binding to proinflammatory chemokines and inhibiting inflammatory cell recruitment. J Immunol 186:3693-3700.
- Gutiérrez-Venegas G, Luna OA, Arreguín-Cano JA, Hernández-Bermúdez C (2014) Myricetin blocks lipoteichoic acid-induced COX-2 expression in human gingival fibroblasts. Cell Mol Biol Lett 19:126-139.
- Kim BJ (2013) Involvement of melastatin type transient receptor potential 7 channels in ginsenoside Rd-induced apoptosis in gastric and breast cancer cells. J Ginseng Res 37:201.
- Pokharel YR, Kim ND, Han HK, Oh WK, Kang KW (2010) Increased ubiquitination of multidrug resistance 1 by ginsenoside Rd. Nutr Cancer 62:252-259.
- Lee SY, Kim GT, Roh SH, Song JS, Kim HJ, et al. (2009) Proteome changes related to the anti-cancer activity of HT29 cells by the treatment of ginsenoside Rd. Int J Pharm Sci 64:242-247.