Keywords
2-deoxy-scyllo-inosose, ribostamycin, Streptomyces ribosidificus NRRL B-11466, gene duplication, ribC, parN.
Introduction
Ribostamycin (RIB) is a core compound and is regarded as an evolutionary ancestor in the neomycin (NEO) family of 2-deoxystreptamine-containing aminocyclitol aminoglycoside antibiotics (2DOS-ACAGAs); as it is a common precursor of all other family members including neomycin, lividomycin (LIV), paromomycin (PAR). The ribostamycin biosynthetic gene cluster has been cloned from S. ribosidificus by two independent research groups [1, 2]. The putative biosynthetic pathway to ribostamycin and related antibiotics was full described [1]. The original postulate that RIB is the precursor to all NEOlike ACAGAs led us to expect that the rib-cluster would be deficient in a glycosyltransferase gene for the third glycosylation (2nd hexosaminyltransfer step). This postulate also established the expectation that this rib-cluster would perhaps lack other genes needed to complete the subsequent steps in the modification pathway, which in turn would lead to the pseudotetrasaccharidic members of the neomycin family.
Therefore, it was surprising to find that the neo- and ribgene clusters are practically identical in gene number and order (Figure 1).
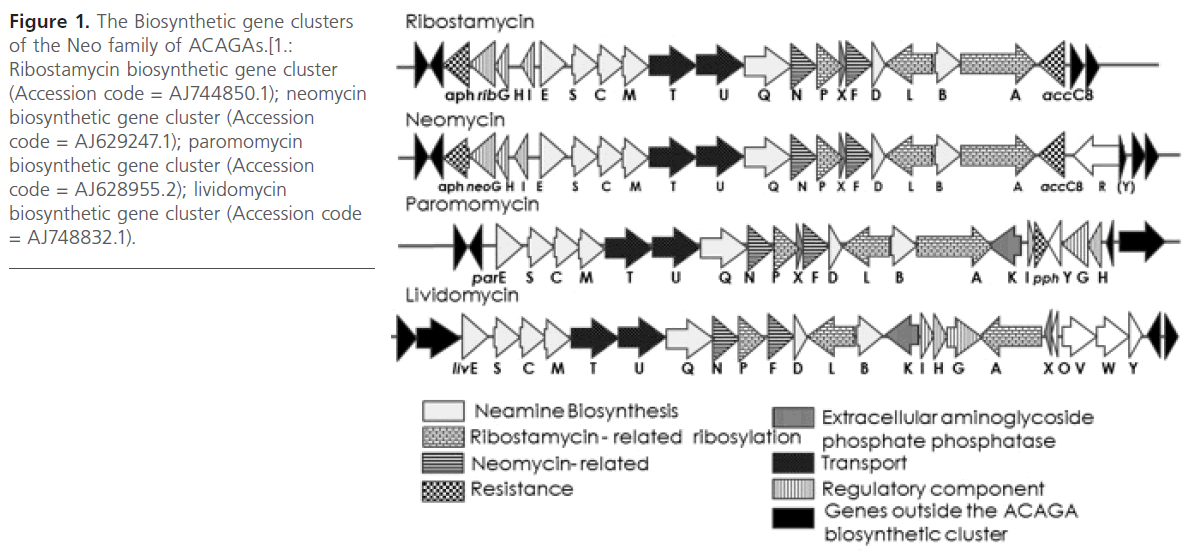
Figure 1: The Biosynthetic gene clusters of the Neo family of ACAGAs.[1.: Ribostamycin biosynthetic gene cluster (Accession code = AJ744850.1); neomycin biosynthetic gene cluster (Accession code = AJ629247.1); paromomycin biosynthetic gene cluster (Accession code = AJ628955.2); lividomycin biosynthetic gene cluster (Accession code = AJ748832.1).
Both gene clusters comprise 20 genes and are flanked by the two resistance genes encoding ACAGA modifying enzymes aminoglycoside phosphotransferase APH(3′) (aphA, rph) and aminoglycoside acetyltransferase AAC(3) (aacC ). The remaining 18 genes should be sufficient to express the functional NEO pathway, in which RIB is an intermediate.
Genes “C,S,E,M,D” and “Q,B” were shown to be involved in paromamine biosynthesis [3, 4, 5] and in the 6′-/6′′′-transamination of hexosamine units [6], respectively. Therefore, they are also conserved in the gene clusters for all other 2-DOS/paromamine-ACAGAs, such as the kanamycin/gentamicin and fortamicin/istamycin families. Of the remaining 11 rib-genes, those conserved in all five clusters for NEO-type ACAGAs (including butirosin) are involved in the remaining steps common in all pathways and which constitute 5 -ribosyltransfer to the 2DOS unit. These are the “L, P” proteins and putatively also the “A” protein [3, 7, 8]. The “G, H, I” genes are postulated to encode a sensor/response regulator system, and the “T, U” genes are postulated to encode a drug exporter belonging to the ABC transporter superfamily. Thus, three genes remain to differentiate the close relatives of the NEO-family pathways (PAR, LIV, NEO, RIB); these are the genes “F, N, X”. The encoded “F” protein was found to be involved in the glycosylation of RIB to give NEO [5]; surprisingly, the ribF gene does not show any indication for being nonfunctional or not being expressed. Therefore, the reason for the absence of the fourth sugar moiety from RIB has to be sought elsewhere. The “X” (possibly a regulatory protein) proteins do not show any evidence for being different between the NEO- and RIB-biosynthetic tools. Therefore, only the “N” proteins, being members of the Fe–S clustercontaining “radical SAM” oxidoreductases (suggesting a biosynthetic function; e.g., for 5′′′-epimerization), remain for differentiating the NEO- and RIB-pathways from each other. Following the analysis of the biosynthetic gene clusters of the NEO family ACAGAs, the aim was to study the two genes ribC and ribN.
ribC was chosen as it is homologous to btrC and neoC which encode 2-deoxy-scyllo-inosose synthase (DOIS); the enzyme which catalyzes the conversion of Glucose-6-phosphate (G- 6-P) to 2-deoxy-scyllo-inosose (DOI) [9, 10]. This is the first step in the biosynthetic pathway of all classes 2DOS ACAGAs. DOI is a unique cyclitol found only in these antibiotics and thus, a typical product of microbial secondary metabolism [11]. Therefore, DOIS catalyzes a key and critical step linking between primary and secondary metabolism in the cell. And since primary metabolites are abundant in the cell, this step may be rate limiting in the biosynthetic process. Consequently, it was proposed that homologous ribC expression in S. ribosidificus (i.e, gene duplication) may increase the antibiotic production. Homologs of ribC are conserved in the biosynthetic gene clusters of all 2-DOS-ACAGAs as well as Isatmycins [1, 12]. So if this proposal was proven to be true, this may be of significant effect on the production of a wide range (almost all clinically used) of ACAGAs which are mainly produced through fermentation.
The cloning of biosynthetic genes is a bottleneck for studying biosynthetic pathways or for their modulations [13]. Therefore, to test the two postulates (ribC gene duplication and ribN effect on production) and to heterologously express RibC, the aim was to clone the two genes ribC and parN from their corresponding strain.
Materials & Methods
Strains and culture conditions
The strains used in this study are listed in Table 1. Streptomyces strains were grown in ISP medium 2 (Bacto yeast extract, 4; malt extract, 5; glucose, 2g/l) and Tryptone soya broth (TSB) (Pancreatic digest of casein, 15; Enzymatic digest of soya bean, 5; Sodium chloride 5g/l). E. coli strains were grown on LB medium (1% tryptone, 0.5% yeast extract, 1% NaCl) supplied 100 μg/ml ampicillin (Sigma) when required. E. coli DH5α was used as the general cloning host. E. coli JM109 (DE3) was the host used for heterologous expression. E. coli ET12567 was used for the preparation of demethylated DNA for transformation into Streptomyces.
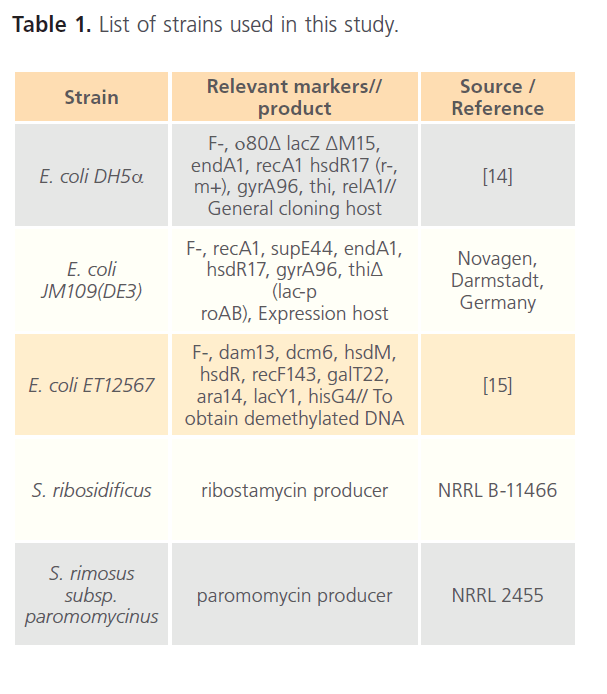
Table 1: List of strains used in this study.
DNA isolation and manipulations
Genomic DNA extractions from S. ribosidificus and S. rimosus were done according to Kieser et al., 2000 [16] based on the method developed by Pospiech and Neumann, 1995 [17]. Cloning, preparation and transformation of competent E. coli cells and in vitro DNA manipulations were carried out according the standard protocols [18].
PCR amplification
The ribC gene was amplified by polymerase chain reaction (PCR) using S. ribosidificus genomic DNA as a template, and the primers: RC-F (5′-AGGGCATATGCAGGTCACGC-3′) and RC-R (5′-CCCGTGCGGATCCACCGGCTA-3′). Likewise the gene parN was PCR amplified using S. rimosus genomic DNA as a template, and the primers PN-F (5′ CACCCCGCATATGACCACC- 3′) and PN-R (5′-ACGGGCGTACCGGATCCGTCCGT-3′). The PCR was performed in thermal cycler, Biocycler TC-S (Boeco, Germany); using 2.5 U Go Taq® Flexi DNA polymerase (Promega). The reaction mixtures (50 μl) contained 250 ng of genomic DNA, 1μM of each primer, 200 μM of each dNTP, 3 mM MgCl2, and 1X Taq buffer. The following conditions were used for the PCR reaction: the enzyme was added after an initial denaturation for 1 minute at 98°C, followed by 30 cycles [98°C for 45 seconds, (66°C ribC; 65°C parN) for 45 seconds, 72°C for 1 minute] and final elongation at 72°C for 5 minutes.
Plasmid construction
The PCR-amplifled products, ribC and parN, were double digested using Fast digest® NdeI and BamHI (Fermentas), and were cloned into the appropriate site of pUCPU21, and a resulting plasmids (pUCRC and pUCPN, respectively) were isolated. After DNA sequencing, the insert DNA (ribC) was isolated and purified after digestion with NdeI/BamHI, and was again ligated into the NdeI-BamHI sites of PET16b and pUWL201PW generating the recombinant plasmids pETRC and pUWRC, respectively. Likewise, the insert DNA in pUCPN (parN) was isolated, double digested (NdeI/BamHI), purified and ligated into the NdeI-BamHI sites of pUWL201PW generating the recombinant plasmids pUWPN. Moreover, the DNA inserts of the respective recombinant plasmids were also verified using DNA sequencing. Table 2 shows the plasmids used in this study.
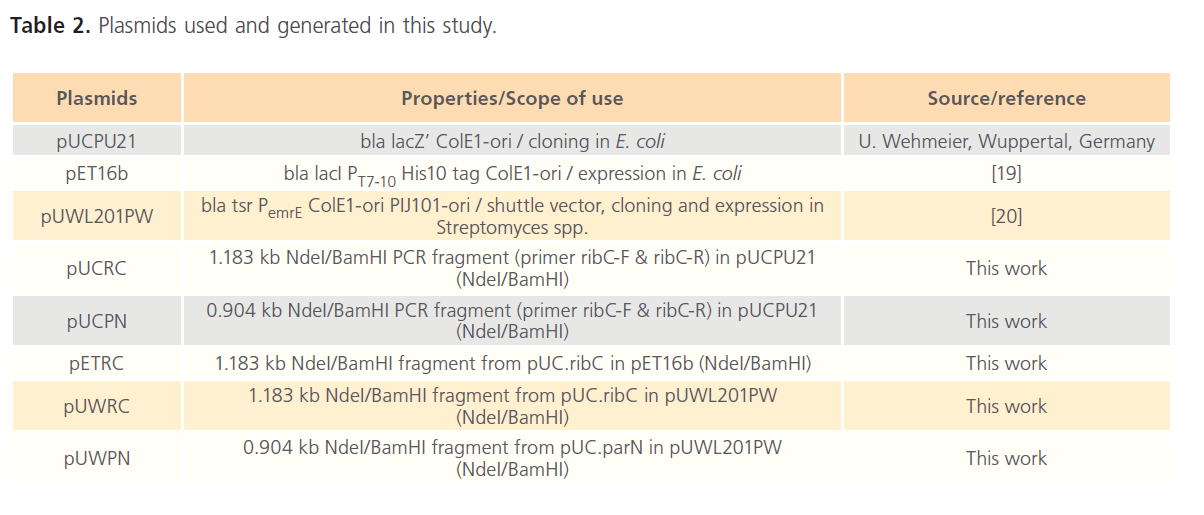
Table 2: Plasmids used and generated in this study.
Overexpression of the ribC gene in E. coli
The recombinant plasmid, pETRC, was transformed into E. coli JM109 (DE3). The preculture of E. coli JM109(DE3)/pETRC was grown in LB medium (100μg/ml ampicillin) overnight at 37°C/300 rpm (xg= 1.12 x R x (RPM/1000)² to saturation. The preculture was used to inoculate a fresh LB/amp growth culture medium (1/100 of volume). The growth culture was grown at 37°C/300 rpm until the OD550 reached 0.5, at which point IPTG was added (final concentration of 1mM) and the incubation was continued at 30°C/55 rpm for 12 hours. About 1 ml aliquots were taken at zero, 1 hr, 2 hr, 4 hr and 12 hr after IPTG induction. For each aliquot, the cells were harvested by centrifugation (13,000 rpm/2-3minutes, 4°C), washed twice with ice-cold 25 mM Tris-HCl pH 7.5, and resuspended in 6 ml cell lysis buffer (25 mM Tris-HCl, pH 7.5; 1 mM Dithiothreitol; 3 mM β-mercaptoethanol; 10 mM MgCl2) per gram E. coli cells. Cells were disrupted by sonication on ice (3 times, each for 45 sec with 30 sec intervals at 60 watt). The cell-free extract was clarified by centrifugation 13000 rpm/15 min at 4oC. Aliquots of the cell-free extracts were run on a 12% SDS-PAGE.
Assay of the expressed RibC protein
The activity of the 2-deoxy-scyllo-inosose synthase (RibC) was determined according to the protocol developed for the AcbC enzyme assay described by Stratmann et al., 1999 [21] with minor modifications. The enzyme assay was performed at 30oC for 6 - 12 hr. The assay was done in a 100 μl reaction mixture containing 12mM G-6-P, 2.5 mM NAD, 4 mM NaF, 0.08 mM CoCl2, 20 mM phosphate buffer (pH 7.5) and 30 μl of the soluble fraction of the cell free extract of RibC harvested 4 and 12 hr after IPTG induction. After incubation the reaction was stopped by heating at 95-100oC for 5 min and centrifuged at 13,000 rpm for 2-3 min. 4-6 μl of the reaction solution was analyzed by TLC and detected with cer reagent. Standard 2-deoxy-scyllo-inosose was kindly provided by Dr. Udo Wehmeier, Wuppertal, Germany.
Transformation of ribC and parN into S. ribosidificus
S. ribosidificus protoplasts were prepared according to Sambrook and Russell, 2001 [18] originally based on the work of Okanishi et al., 1974 [22] which was optimized for high transformation frequency by Bibb et al.,1978 [23]. Mycelia were grown at 28oC/200 rpm for 36-40 hr in 25 ml TSB supplemented with 5% PEG 6000, 0.5% glycine and 5mM MgCl2 with a stainless steel spring. The mycelia were pelleted by centrifugation at 3000 rpm/10 min, washed twice with 10.3% sucrose solution and resuspended in 4 ml L-buffer or lysozyme solution (lysozyme (MP biomedicals) dissolved in Pbuffer at a concentration of 1 and 2 mg/ml) and incubated at 30-37oC for 15-60 min till protoplast formation was evident. 5 ml P-buffer (sucrose, 103; K2SO4, 0.25; MgCl2·6H2O, 2.02 g/800 ml, trace elements solution 2 ml/800 ml; dispense in 80 ml aliquots and autoclave; Before use add to each aliquot 0.5% KH2PO4, 1ml; 3.68% CaCl2.2H2O, 10ml; 5.73% TES buffer pH7.2, 10 ml. Trace elements solution: ZnCl2, 40 mg; FeCl3.6H2O, 10; CaCl2 2H2O, 10; MnCl2.4H2O, 10; Na2B4O7.10H2O, 10; (NH4)6Mo7O24.4H2O, 10mg/l) was then added and the protoplast suspension was filtered through sterile cotton wool. The protoplasts were sedimented by centrifugation at 3000 rpm/7 min and resuspended in 1 ml P-buffer. The protoplasts were transformed according to Babcock and Kendrick, 1988 [24]. 5 μl of the demethylated recombinant DNA (isolated from the methylation-deficient strain E. coli ET12567) was added to 50 μl freshly prepared protoplast suspension and mixed immediately by finger tapping. 200 μl T-buffer (or 25% PEG 1500 in P-buffer) was immediately added and mixed gently by pipetting up and down several times. After the addition of 1.3 ml P-buffer, the cells were plated out onto pre-dried protoplast regeneration medium (SpMR). The cells were allowed to recover by an overnight incubation at 30oC, after which they were flooded with 1ml of dilute thiostrepton (MP biomedicals). Cells were allowed to grow under the appropriate selection pressure (50 μg/ml thiostrepton). Plates were checked for transformants after 2-5 days.
Results & Discussion
For an elucidation of the genetics and the biosynthetic pathways for the production of the ribostamycin and related antibiotics, the biosynthetic gene clusters of the respective antibiotics were fully isolated, sequenced, analyzed and annotated [1, 4, 25]. Analysis of the biosynthetic gene clusters for the major 2DOS-ACAGAs revealed greater similarities in the both gene/enzyme sequences and arrangement particularly to those involved in the biosynthesis of 2DOS moiety, the basic aglycone unit [1, 4, 25]. The similarity of the chemical structures of the regarded antibiotics was mirrored by their greater similarity in the respective gene clusters, gene content and sequence similarity in the individual genes/enzymes [1]. Basically, the biosynthesis of these antibiotics in their producers exhibits a common biosynthetic pathway which is then followed by further unique biosynthetic steps that will lead to the different members of this subclass [1]. Furthermore, both the rib- and neo-clusters are highly conserved and the only major difference found was that in the rib-cluster, a natural frame shift mutation created by two compensating frame shifts (+1 bp in position 544 and -1 in position 771) was seen in the ribN open reading frame (ORF) (encoding a putative 5’’’-epimerase; Figure 2) [1].

Figure 2: (A) Alignment of RibN and its homologous proteins (accession code = CAG34032) and the homologous proteins: ParN (accession code = CAF32379), livN (accession code = CAG38702and NeoN (accession code = CAF33317) and (B) their phylogram.
Alignment of RibN and homologous proteins
The RibN protein shows a remarkable difference in its primary structure relative to those of the NeoN, ParN, and LivN proteins: A stretch of nonconserved amino acid sequence is observed at the positions from 185 to 261 in the RibN amino acid sequence as determined by multiple alignment sequences of the respective proteins (Figure 2). Moreover, this deviating sequence is created by two compensating frame-shifts (+1/−1) in the nucleotide sequence of ribN between which the sequence shows the same degree of conservation as that of the neoN gene as outside this region [1]. These results can be interpreted by either one or a combination of the following possibilities: (i) the RibF protein has lost its function; however, its production cannot be excluded because there are no features to be seen in the DNA sequence which would indicate its exclusion from transcription or translation; (ii) the natural frame-shifts occurring in the ribN ORF lead to an inactive RibN protein which could negatively affect this glycosylation process (RibN could be involved in other, as yet, unknown accompanying function(s) affecting the fidelity of the third glycosylation step); (iii) the trisaccharidic nature of ribostamycin could be a favorite substrate for the exporter system (RibT/U) and therefore, compete with the third glycosylation (RibF) which leads to pumping of ribostamycin outside the cell before being further glycosylated [1]. Therefore, The gene parN (without frame-shift mutation) from Streptomyces rimosus subsp. paromomycinus NRRL 2455 was also amplified using PCR and cloned into the cloning pUCPU21 and shuttle pUWL201PW vectors in an attempt for transformation and heterologous expression in S. ribosidificus and to study its role in ribostamycin production.
Construction of recombinant plasmids
Both ribC and parN were amplified using PCR from the genomic DNA of S. ribosidificus and S. rimosus, respectively. The forward primers were designed to introduce an NdeI site (underlined) in place of the natural start codon and for the ability to create a start codon fusion of ribC and parN downstream from the ribosome-binding-site of cloning pUCPU21, expression pET16b, and shuttle pUWL201PW vectors. On the other hand, the reverse primers were designed for the introduction of an alternate restriction site located immediately downstream of the natural stop codon in order to allow orientated cloning into the cloning and expression vectors. The recombinant plasmids pUCRC, pUCPN, pETRC, pUWRC, and pUWPN constructed in this work before and after NdeI/ BamHI digestion were tested and analysed using agarose gel electrophoresis (Figure 3, Figure 4).
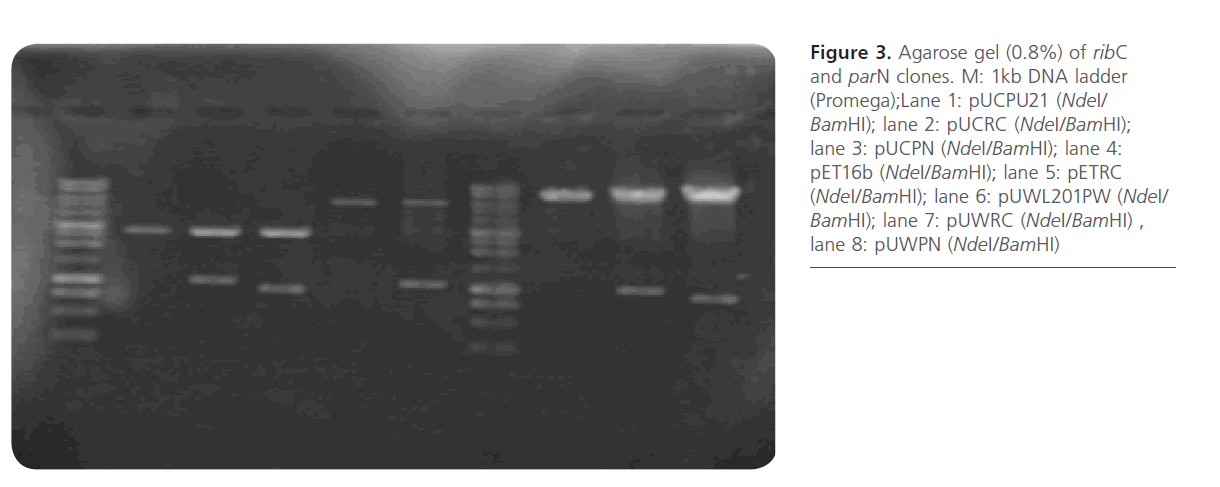
Figure 3: Agarose gel (0.8%) of ribC and parN clones. M: 1kb DNA ladder (Promega);Lane 1: pUCPU21 (NdeI/ BamHI); lane 2: pUCRC (NdeI/BamHI); lane 3: pUCPN (NdeI/BamHI); lane 4: pET16b (NdeI/BamHI); lane 5: pETRC (NdeI/BamHI); lane 6: pUWL201PW (NdeI/ BamHI); lane 7: pUWRC (NdeI/BamHI) , lane 8: pUWPN (NdeI/BamHI)
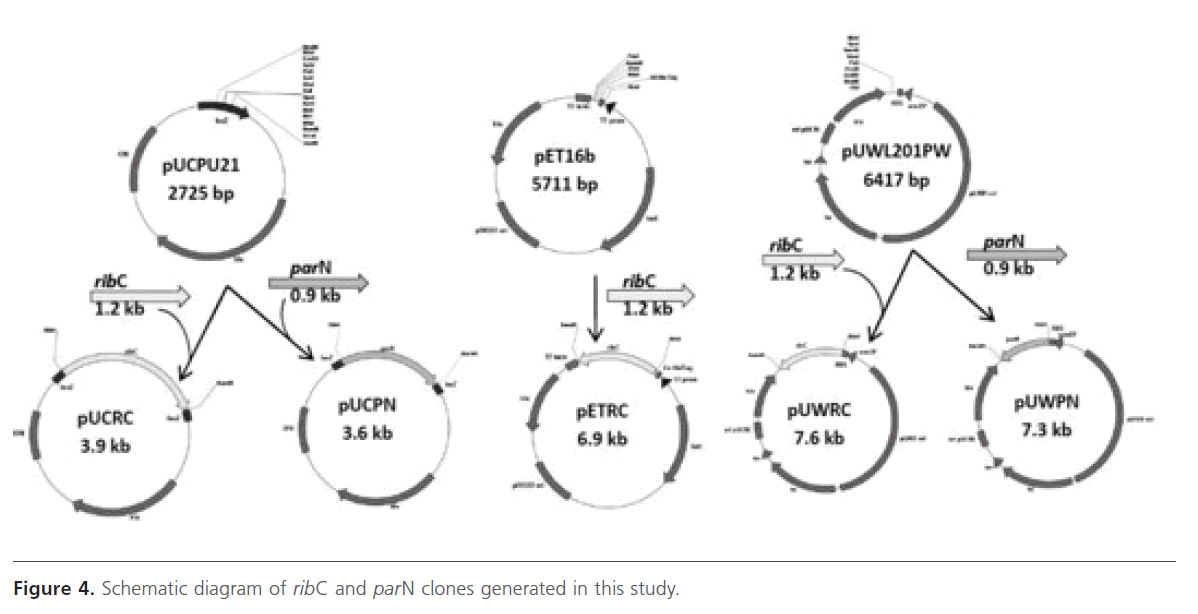
Figure 4: Schematic diagram of ribC and parN clones generated in this study.
Expression of ribC in E. coli JM109(DE3)
As shown in Figure 5, RibC was overproduced as soluble N-terminal His-tagged protein in E. coli JM109 (DE3) as determined by SDS-PAGE. An additional band of about 42 kDa corresponding to the expected molecular mass of the Histagged RibC protein was observed in the soluble fraction of the cell-free extracts. Maximal expression of RibC was 4h after IPTG induction at 30oC/55 rpm. In this work, the soluble N-terminally His-tagged RibC protein was only achieved via heterologous expression in the E. coli strain.
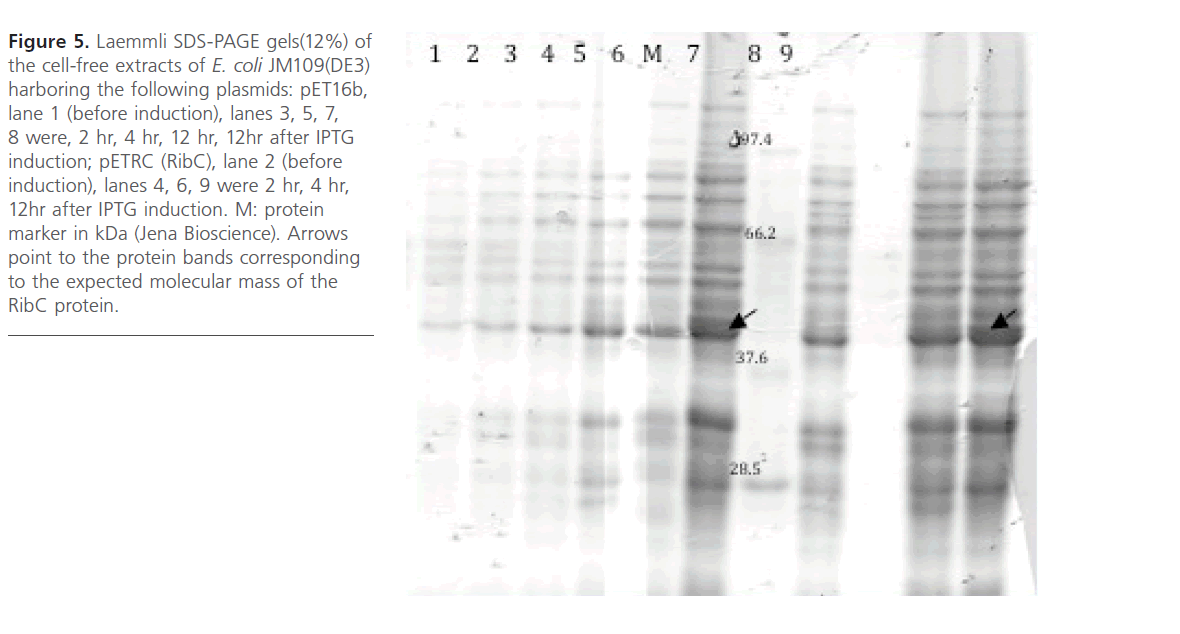
Figure 5: Laemmli SDS-PAGE gels(12%) of the cell-free extracts of E. coli JM109(DE3) harboring the following plasmids: pET16b, lane 1 (before induction), lanes 3, 5, 7, 8 were, 2 hr, 4 hr, 12 hr, 12hr after IPTG induction; pETRC (RibC), lane 2 (before induction), lanes 4, 6, 9 were 2 hr, 4 hr, 12hr after IPTG induction. M: protein marker in kDa (Jena Bioscience). Arrows point to the protein bands corresponding to the expected molecular mass of the RibC protein.
RibC catalyzed the formation of DOI
The soluble fractions containing His-tagged RibC protein [JM109(DE3)/pETRC] 4 and 12 hr after IPTG induction were used for analysis. Conversion of G-6-P into 2-deoxy-scylloinosose was detected by TLC using cer-reagent. A new spot on the TLC sheet with an Rf-value corresponding to the migration of the standard 2-deoxy-scyllo-inosose was observed in the enzymatic reaction containing the soluble 4 hr Histagged RibC protein and to a lower extent in that containing the soluble 12 hr protein. In control reactions, containing the empty vector (pET16b) or containing no G-6-P. no conversion of G-6-P to 2-deoxy-scyllo-inosose was observed (Figure 6).

Figure 6: TLC analysis of RibC protein catalyzed conversion of G-6-P into 2-deoxy-scyllo-inosose. Lane 1: standard Glucose-6- phosphate; lane 2: standard 2-deoxy-scyllo-inosose; lane 3: pETRC 4 hr; lane4: pET16b 4hr; lane5: pETRC 12 hr; lane 6: pET16b 12hr; lane 7: PETRC 4hr, No G-6-P; lane 8: pET16b 4hr, No G-6-P.
Role of RibC in ribostamycin biosynthesis
DOI synthase switches the flux of G-6-P from the intercellular primary metabolite pool to a secondary metabolite biosynthetic route through the synthesis of a non-aminogenous cyclitol, DOI [26]. Thus, it is a key enzymatic step in the biosynthetic pathway of 2-DOS-containing ACAGAs. RibC was verified biochemically to be involved in the conversion of G-6-P into 2-deoxy-scyllo-inosose. In this context, a promising strategy that could lead to the higher production of ribostamycin, and probably all 2-DOS-containing ACAGAs, would be the construction of a cassette for the DOI synthase gene and its subsequent transformation into S. ribosidificus or other producing strains. Also, DOI is valuable as a starting material for the benzene-free synthesis of catechol and other benzenoids [27, 28, 29]. Protoplasts of S. ribosidificus were successfully transformed with pUWRC (PUW201PW + ribC gene, 1.2 kb) for the purpose of ribC gene duplication (one cope was located in the chromosome and other copy was on the recombinant shuttle vector, pUWRC). Results showed that the pUWRC transformant was able to grow on thiostrepton (50μg/ml)-containing SpMR plates, while the wild S. ribosidificus was unable to grow under these conditions. The pUWRC transformants were selected to be tested for their ribostamycin production in comparison to the wild strain.
Conclusion and prospective of this work
The detection of DOI synthase activity in the cell-free extract of E. coli JM109 (DE3)/pETRC represents the first report of expression of functional 2-deoxy-scyllo-inosose from S. ribosidificus in E. coli. DOI is valuable as a starting material for the benzene-free synthesis of catechol and other benzenoids. Both ribC and parN were successfully cloned into pUWL201PW shuttle vector producing pUWRC and pUWPN, respectively. The resulted recombinant plasmids were transformed into S. ribosidificus for the purpose of gene duplication and studying their influence on ribostamycin production.
Acknowledgement
We acknowledge Department of Microbiology and Immunology, Faculty of Pharmacy, Ain Shams University for their support and providing equipment facilities.
82
References
- Piepersberg, W., Aboshanab, KM., Schmidt-Beißner, H., Wehmeier, UF. In Aminoglycoside antibiotics (Arya D P ed) John Wiley & Sons. Inc pp 15-118, 2007.
- Subba, B., Kharel, MK., Lee, HC., Liou, K., Kim, BG. et al. The ribostamycin biosynthetic gene cluster in Streptomyces ribosidificus: comparison with butirosin biosynthesis. Molecules & cells 2005; 20: 90-96.
- Llewellyn, NM., Spencer, JB. Biosynthesis of 2-deoxystreptamine-containing aminoglycoside antibiotics. Nat Prod Rep. 2006; 23: 864-874.
- Kudo, F., Yamamoto, Y., Yokoyama, K., Eguchi, T., Kakinuma, K. Biosynthesis of 2-deoxystreptamine by three crucial enzymes in Streptomyces fradiae NBRC 12773. J Antibiot. 2005; 58: 766-774.
- Yokoyama, K., Yamamoto, Y., Kudo, F., Eguchi, T. Involvement of two distinct N-acetylglucosaminyltransferases and a dual-function deacetylase in neomycin biosynthesis Chembiochem. Euro J chemi biology 2008; 9: 865-869.
- Huang, F., Spiteller, D., Koorbanally, NA., Li, Y., Llewellyn, NM. et al. Elaboration of neosamine rings in the biosynthesis of neomycin and butirosin Chembiochem. Euro J chemi biology 2007; 8: 283-288.
- Kudo, F., Fujii, T., Kinoshita, S., Eguchi, T. Unique O-ribosylation in the biosynthesis of butirosin. Bioorg Med Chem. 2007; 15: 4360-4368.
- Huang, F., Haydock, SF., Mironenko, T., Spiteller, D., Li, Y. et al. The neomycin biosynthetic gene cluster of Streptomyces fradiae NCIMB 8233: characterisation of an aminotransferase involved in the formation of 2-deoxystreptamine. Organic & biomolecular chemistry 2005; 3: 1410-1418.
- Yamauchi, N., Kakinuma, K. Confirmation of in vitro synthesis of 2-deoxy-scyllo-inosose the earliest intermediate in the biosynthesis of 2-deoxystreptamine using cell free preparations of Streptomyces fradiae. J Antibiot 1992; 45: 774-780.
- Kudo, F., Hosomi, Y., Tamegai, H., Kakinuma, K. Purification and characterization of 2-deoxy-scyllo-inosose synthase derived from Bacillus circulans A crucial carbocyclization enzyme in the biosynthesisof 2-deoxystreptamine-containing aminoglycoside antibiotics. J Antibiot. 1999; 52: 81-88.
- Yamauchi, N., Kakinuma, K. Enzymic carbocycle formation in microbial secondary metabolism The mechanism of the 2-deoxy-scyllo-inosose synthase reaction as a crucial step in the 2-deoxystreptamine biosynthesis in Streptomyces fradiae. J Org Chem. 1995; 60: 5614-5619.
- Kudo, F., Eguchi, T. Biosynthetic genes for aminoglycoside antibiotics. J Antibiot 2009; 62: 471-481.
- Kurumbang, NP., Park, JW., Yoon, YJ., Liou, K., Sohng, JK. Heterologous production of ribostamycin derivatives in engineered Escherichia coli. Res Microbiol. 2010; 161: 526-533.
- Hanahan, D. Studies on transformation of Escherichia coli with plasmids. J Mol Biology 1983; 166: 557-580.
- MacNeil, DJ., Gewain, KM., Ruby, CL., Dezeny, G., Gibbons, PH., MacNeil, T. Analysis of Streptomyces avermitilis genes required for avermectin biosynthesis utilizing a novel integration vector. Gene 1992; 111: 61-68.
- Kieser, T., Bibb, MJ., Buttner, MJ., Chater, KF., Hopwood, DA. Practical Streptomyces genetics, p. 613, 2000.
- Pospiech, A., Neumann, B. A versatile quick-prep of genomic DNA from Gram-positive bacteria. Trends Genet 1995; 11: 217-218.
- Sambrook, J., Russell, DW. Molecular cloning: a laboratory manual 3rd ed Cold Spring Harbor Laboratory, Press Cold Spring Harbor NY, 2000.
- Studier, FW., Rosenberg, AH., Dunn, JJ., Dubendorff, JW. In Methods in Enzymology vol. 185 (David V G ed) Academic Press, p. 60-89, 1990.
- Doumith, M., Weingarten, P., Wehmeier, UF., Salah-Bey, K., Benhamou, B. et al. Analysis of genes involved in 6-deoxyhexose biosynthesis and transfer in Saccharopolyspora erythraea. Mol Gen Genet. 2000; 264: 477-485.
- Stratmann, A., Mahmud, T., Lee, S., Distler, J., Floss, HG. et al. The AcbC protein from Actinoplanes species is a C7-cyclitol synthase related to 3-dehydroquinate synthases and is involved in the biosynthesis of the alpha-glucosidase inhibitor acarbose. J Biol Chem. 1990; 274: 10889-10896.
- Okanishi, M., Suzuki, K., Umezawa, H. Formation and reversion of Streptomycete protoplasts: Cultural condition and morphological study. J Gen Microbiol. 1974; 80: 389-400.
- Bibb, MJ., Ward, JM., Hopwood, DA. Transformation of plasmid DNA into Streptomyces at high frequency. Nature 1978; 274: 398-400.
- Babcock, MJ., Kendrick, KE. Cloning of DNA involved in sporulation of Streptomyces griseus. J Bacteriol. 1988; 170: 2802-2808.
- Ota, Y., Tamegai, H., Kudo, F., Kuriki, H., Koike-Takeshita, A. et al. Butirosin-biosynthetic gene cluster from Bacillus circulans. J Antibiot. 2000; 53: 1158-1167.
- Iwase, N., Kudo, F., Yamauchi, N., Kakinuma, K. Substrate specificity of 2-deoxy-scyllo-inosose synthase the starter enzyme for 2-deoxystreptamine biosynthesis toward deoxyglucose-6-phosphates and proposed mechanism. Biosci Biotechnol Biochem. 1998; 62: 2396-2407.
- Kakinuma, K., Nango, E., Kudo, F., Matsushima, Y., Eguchi, T. An expeditious chemo-enzymatic route from glucose to catechol by the use of 2-deoxy-scyllo-inosose synthase. Tetrahedron Letters 2000; 41: 1935-1938.
- Hansen, CA., Frost, JW. Deoxygenation of polyhydroxybenzenes: An alternative strategy for the benzene-free synthesis of aromatic chemicals. J Am Chem Soc. 2002; 124: 5926-5927.
- Nango, E., Kumasaka, T., Hirayama, T., Tanaka, N., Eguchi, T. Structure of 2-deoxy-scyllo-inosose synthase a key enzyme in the biosynthesis of 2-deoxystreptamine-containing aminoglycoside antibiotics in complex with a mechanism-based inhibitor and NAD+. Proteins 2008; 70: 517-527.













