INTRODUCTION
|
| |
| Locked nucleic acids have efficiently suppressed telomerase activity in cells (a critical task for cancer fighting on a genetic level) and have prevented Tatdependent transcription, and thus the spread of cancers. Locked nucleic acids have been used to modify DNAzymes to create LNAzymes, and these have been shown highly useful in efficiently cleaving highly structured RNA[1]. |
| |
| Locked nucleic acid (LNA) is a novel class of DNA analogues and was invented by Professor Jesper Wengel,University of Copenhagen & Dr.Poul Nielsen,Odense University in 1996[3]. |
| |
| The LNA monomer is a bicyclic compound in which the 2’ & 4’ positions of the furanose ring are linked by an O-methylene (oxy-LNA),S-methylene (thio-LNA) or NH-methylene moiety (amino LNA). This linkage restricts the conformational freedom of the furanose ring and hence the name a locked nucleic acid. |
| |
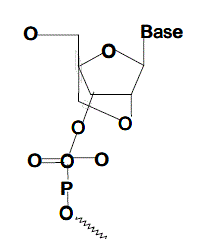 |
| |
| Introduction of LNA monomers into DNA or RNA oligomers dramatically increases their affinity for complementary DNA or RNA ( measured as the thermal stability,of the duplex, Tm). A DNA duplex ( 5’-GTGATATGC-3’:5’-GCATATCAC-3’) have a Tm of 28 C. in 100 mM Nacl, where as the Tm of the similar LNA:DNA heteroduplex is 64 C. In general, increase in affinity ranges from 3 to 8 C. per LNA monomer depending on the sequence and number of LNA monomers in the oligo. LNA modified oligomers have been found to obey the Watson-Crick hydrogen bonding rules and to display exellent target specificity[2]. |
| |
| LNA Monomer Structures[4] are given below :- |
| |
 |
| |
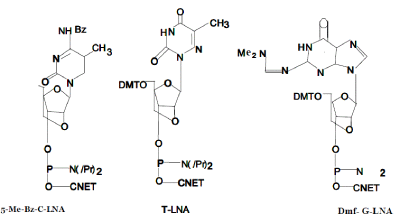 |
| |
|
5-Me-Bz-C-LNA
|
| |
| The extraordinary high Tm of LNA and its biological stability make it ideal for antisense drug development or as antisense molecule and the potential of LNA to inhibit the growth of Cancer cells. |
| |
AS CANCER THERAPEUTIC AGENT
|
| |
| The antisense (AS) technique holds the potential to specifically down-regulate the expression of any selected gene. Thus, in principle it should be possible to design an antisense drug to target any mRNA encoded by the human genome and potentially have specific treatment for any disease caused by overexpression of a single (or a few) gene(s). |
| |
| LNA is a novel class of nucleic acid analogues, which exhibits unprecedented thermal stability towards complementary LNA, DNA and RNA. Furthermore, LNA obeys Watson-Crick base pairing rules, is stable in serum and can be taken up by mammalian cells. In vivo, LNA displays low toxicity and an LNA antisense oligonucleotide (AON) has been demonstrated to inhibit tumour growth in a murine xenograft model. All together, these characteristics of LNA make it a very promising new player in the antisense field of research. |
| |
| LNA oligonucleotides are more potent than the currently used phosphorothioate AONs, LNA gapmer AONs are highly efficient antisense molecules. LNA can be applied as antisense molecule or oligonucleotide and developed as Cancer therapeutic drugs [5]. |
| |
| The use of a complementary sequence can inhibit the expression of aspecific mRNA, inducing a blockade in the transfer of genetic information from DNA to protein. This principle was proposed in 1967 by Belikova and colleagues, who suggested that RNA sequences serve as endogenous inhibitors of gene expression in prokaryotes[6]. Ten years later, Paterson and colleagues found that exogenous, singlestranded nucleic acids inhibit RNA translation in cell-free systems[7]. In 1978, Zamecnik and Stephenson showed the potential of oligodeoxynucleotides to act as antisense agents that inhibit viral replication in cell cultures [8]. |
| In the last two decades, with the advent of automated DNA synthesis, advances in the field of nucleic-acid chemistry, progress in the possibility of rational design and the accumulation of different genome sequences have led to the use of short fragments of oligonucleotides, either as therapeutic agents or as tools to study gene function. The development of antisense oligonucleotide technologies as therapeutic agents has led to Food and Drug Administration approval for the commercialisation of the first (and to date only) antisense oligonucleotide, Vitravene, for the treatment of cytomegalovirus-induced retinitis in patients affected by AIDS, and to numerous clinical trials of therapeutic Oligonucleotides[9,10]. |
| |
MECHANISM OF ACTION OF ANTISENSE OLIGONUCLEOTIDES
|
| |
| Antisense oligonucleotides usually consist of 15-20 nucleotides, which are complementary to their mRNA target.On the basis of the mechanism of action, two classes of antisense oligonucleotides can be devised: (a) the RNase H-dependent oligonucleotides, which induce degradation of mRNA; and (b) the steric-blocker oligonucleotides, which physically prevent or inhibit the progression of splicing or the translational machinery. The majority of antisense oligonucleotides investigated in clinical trials act via RNase H-dependent mechanism. |
| |
TRIPLE HELIX-FORMING
|
| |
OLIGONUCLEOTIDES AND ANTIGENE STRATEGY
|
| |
| Double-stranded DNA is the main form of genetic material storage in living organisms and for this reason represents an attractive target for genetic modification and gene-directed therapy. In contrast to the antisense treatment, direct modification of a target gene renders the results of this treatment irreversible, opening the possibility for the artificial control of gene expression at the whole genome level. |
| |
| Besides natural proteins and peptides, two classes of synthetic substances that recognise double-stranded DNA sequences are known: triple helix-forming oligonucleotides (TFO)[11,12] and oligocarboxamide minor groove binders (MGB)[13,14,15].The recognition of DNA by TFO is based on formation of Hoogsteen (or reverse Hoogsteen) base triplets when polypyrimidine (or polypurine) single strand forms hydrogen bonds with a polypurine tract in a major groove of double-stranded DNA[16,17]. |
| |
| Protonation of cytosine in the polypyrimidine third strand is necessary for hydrogen bond formation; hence the triple helix becomes stable in slightly acidic conditions (pH<6). Oligonucleotides with modified backbones are more and more frequently used for triplex formation.For example, 2’-Omethyloligoribonucleotides form quite stable triple helices with the target DNA having the advantage of a simple synthetic procedure and a high stability toward nuclease degradation[18,19].Very stable triplexes are formed by (a) phosphoramidates,[20,21] where one of the backbone oxygen atoms is replaced by a nitrogen atom, (b) locked nucleic acids (LNA) where 2' and 5'-positions of ribose are linked by a methylene bridge[22-24] and (c) peptide nucleic acids (PNA) that possess nucleic bases and a peptide backbone at the same time[25-28] . |
| |
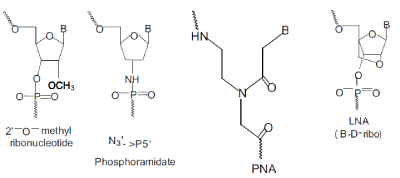 |
| |
| Examples of modified Backbones used to Synthesise new generations of Triple helix- forming Oligonucleotides. |
| |
LOCKED NUCLEIC ACID
|
| |
| A very novel approach to antigene and antisense therapy is represented by the introduction of locked nucleic acids . These compounds have been the subjects of interesting and exhaustive reviews [23,24,27,28]. They were synthesised for the first time in 1998 [30,31-33] and preseuted as sugar ring modifications with respect to the natural nucleic acids. The variation consists in a constriction between O2’ and C4’, by the linkage of the two units owing to a locked C3’-endo sugar conformation. This structure is optimal for RNA because it mimics the natural conformation of ribonucleic acid |
| |
 |
| |
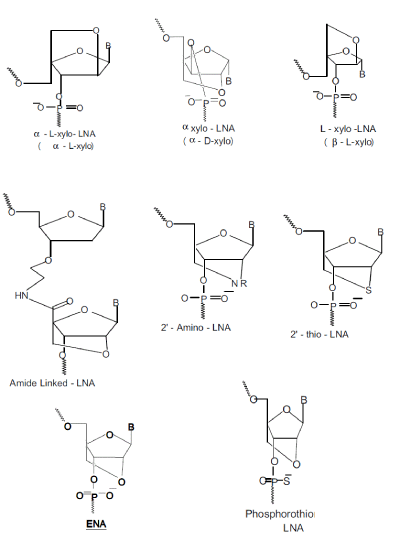 |
| |
|
LNA Isomers and Derivatives
|
| |
| LNAs are easy to handle and offer many advantages. Their introduction on 3’ or 5’ end confers to the oligonucleotides a good resistance to nucleases. Generally they are able to activate RNase H, probably because of the conformational perturbation induced by their presence [34]. The phosphate groups in the structural backbone facilitated their transportation into cell by utilizing traditional cationic vectors and confer good water-solubility to these compounds. LNA units can be incorporated into oligonucleotides to give chimeras following the standard procedures for DNA synthesis. LNAs are able to discriminate a mismatch better than native nucleic acids and they are able to form very stable LNA/DNA or LNA/RNA duplexes. In fact, the presence of LNA into a complementary strand increases the stability of the duplex by +1 - +8OC against DNA and by +2 - +10OC against RNA, for each introduced monomer [30,31,33]. Christensen et al. performed studies on the kinetics and the thermodynamics of duplex formation between LNA and DNA[35]. Wengel and co-workers explain that the stability of the partly modifie LNA/DNA duplex can be due to a decreased loss in entropy and to an increased loss in enthalpy upon duplex formation by organisation of the phosphate backbone, favouring a more efficient stacking of nucleobases [36]. |
| |
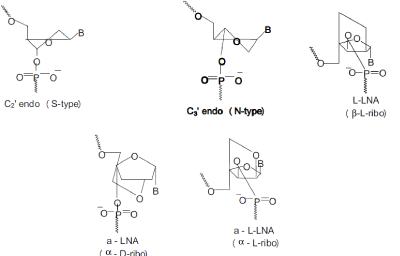
| Different synthetic methodologies have been described to obtain LNAs, their isomers or analogus[37-46] and many efforts are ongoing to introduce further modifications, i.e. the preparation of ENA, 2’-O,4’-C-ethylene-bridged nucleic acids and the corresponding phosphoramidites [47]. All eight possible LNA stereoisomers. have been synthesised and six of them exhibit remarkable binding affinity towards RNA[42] . The O-LNA presents the sugar moiety with O-D-configuration and three different approaches towards thymine and 5-methyl-cytosine monomers have been reported[38] . Håkansson and Wengel described O-L-LNA adenine derivative synthesis and its high affinity hybridisation towards complementary sequences[48] while Nielsen et al. analysed the sugar conformation from NMR coupling constants and CD spectroscopy showing that O-LLNA/ DNA duplex adopts general B-like geometry [49]. |
| |
| 2’-Thio-LNA and phosphorothioate-LNA and methylphosphonate- LNAs[50] have been synthesised. The 2’-Thio-LNA derivatives show a dramatic increase of thermal stability in LNA/DNA or LNA/RNA duplexes (about 5°C for each modification in DNA binding and 8°C in RNA binding). The presence of phosphorothioate- LNAs or methylphosphonate-LNAs is more efficient than the phosphorothioate nucleic acid analogues but less than LNA[39]. Despite this, such modification will play an important role because the presence of one methylphosphonate-LNA in a 9-mer sequence is sufficient to prevent 3-exonucleolytic degradation[50] . Also different amide-linked LNAtype dinucleotides have been prepared and incorporated into oligodeoxynucleotides, with significant increase of thermostability of the studied duplex[51]. |
| |
| Kurreck et al. found that the presence of seven or eight DNA monomers in a DNA/LNA chimera is necessary for a satisfying activation of RNase H, while in 2’-O-methyl gapmers only six natural monomers are required to obtain the same activation. They also demonstrated that three LNAs at each end of the strand increase the stability ten times more than the native oligonucleotide and phosphorothioates or 2'- O-methyl gapmers give a lower stabilisation with respect to LNA[52].Also Keinicke et al. studied LNA and O-L-LNA chimeras, obtaining RNA-selective hybridisation, increasing the binding affinity and the stabilisation toward exonucleases[53] . |
| |
APPLICATIONS OF LNA
|
| |
| Many different applications for these modified oligonucleotides have been demonstrated. The inhibition of gene expression of HIV-1 Tat dependent trans-activation in HeLa cell nuclear extract, where LNA chimeras give 50% of inhibition at concentrations of about 100 nM [54]. |
| |
| To stop RNA function by taking advantage of the RNA tendency to assume different folded states having similar free energy. This technique is called oligonucleotide directed misfolding of RNA (ODMiR), and utilizes exogenous oligonucleotides to stabilise inactive structures. These chimeras inhibit 50% of intron splicing at nanomolar concentrations, inducing folding alteration[55] . |
| |
| Ørum et al. used an ELISA-like assay to detect the factor V Leiden mutation in PCR-amplified genomic DNA using LNA octamer probes and this successful method demonstrated excellent sensitivity and specificity[56] .LNAs have been also used in the detection of a single nucleotide polymorphism in apolipoprotein E, applying this assay on patients with unambiguous results[57] . |
| |
| Vectors containing at each terminus one LNA residue facilitate correction of point and frameshift mutations. The rectification frequency increases progressively with the increase of the number of LNA bases. These results indicate in the possibility of utilising LNA oligonucleotides in direct single nucleotide change reactions in vivo[58] . |
| |
| The LNA strong affinity towards matching DNA and RNA has been exploited to inhibiting human telomerase. LNA oligomers, complementary to the telomerase RNA template, are potent and selective inhibitors of this protein, strongly involved in tumoral processes[59]. |
| |
| Corey and co-workers examined the regulation that controls antisense gene inhibition within cells. The modified oligonucleotides were delivered utilising standard procedure that implies use of ationic lipids. The oligos target the terminus of the 5’-untranslated region inhibiting the luciferase expression, probably by blocking the binding of ribosomes to the transcript[60] . |
| |
| Nuclear transcription factor KB (NF-KB) and other DNAbinding proteins do not have a good binding affinity for LNA oligomers. Crinelli et al. studied the influence of LNA insertion in decoy molecules for NF-OB binding sequence, present in the PRDII domain of interferon B II [61] . If the LNAs are in terminal positions of the utilised strand, these oligonucleotides show an interesting stability toward nucleases. LNA introduction into the NF-KB recognition portion leads to an enhancement of the stability, but with loss of affinity to the target, probably because of small changes in the molecular structure of the KB-binding sequence with a local architecture change of phosphate geometry. The interaction changes depending whether LNAs are used as sense or antisense strand: higher affinity of NF- KB was observed in the former case with respect to the latter. It was assumed that LNA sense strand might effect the conformation of the AT-rich sequences of PRDII in the minor groove; on the contrary, the modified antisense strand might alter the G-C rich sequence of PRDII, which are strongly involved in interacting with the transcription factor. This can lead to a more consistent decrease of affinity for NF-KB for its binding element. |
| |
| The possibility to utilise LNAs as decoy agents and, despite the loss of affinity if the modifications are introduced into NF-OB recognition portion, the decoy molecules have been shown to compete with a DNA probe in nanomolar concentrations[61] |
| |
| Wahlestedt et al. illustrated the first in vivo study with LNA and DNA/LNA antisense oligonucleotide chimeras in 2000[62] and analysed the cellular uptake of an LNA 15-mer in human MCF-7 cells, presenting the same localisation in living and in fixed cells. These modified oligonucleotides, built to knockdown the rat delta opioid receptors, were demonstrated to have good stability in serum and to be able to activate RNase H. Besides, if injected into rat brain parenchyma, they do not elicit toxicity in contrast with phosphorothioate oligonucleotides that induce severe damage in administration tissues. The antisense oligonucleotides were effective in vivo as the blockage of the G-protein coupled opioid receptor was demonstrated by the inactivity of deltorphin II, selective agonist for this receptor, which, after activation, usually induces hypothermia and spinal antinociceptive response in the tail-flick test. |
| |
| Fluiter et al. have recently demonstrated the efficacy of LNAs in the tumour growth inhibition in vivo[63] . As target, they selected the gene codifying for the large subunit of RNA polymerase II (POLR2A). The full LNA oligonucleotide was tested in prostate cancer cells (15PC3). Despite the fact that it was not able to activate RNase H, the oligo reduced the POLR2A protein expression for the two forms of POLR2A (hyper- and hypophosphorylated), with a preference for the former one, without mRNA degradation. |
| |
| Tatiana Da Ros et al evidenced a 40% reduction of protein expression at 50 nM and also a higher LNA concentration into cells with respect to Phosphorothioate oligonucleotides and performed determinations of serum decay and biodistribution of LNA in nude mice and compared the results with those obtained from phosphorothioate analogues. No main differences in biodistribution were observed with the exception of urinary excretion, which is extremely fast and higher for LNA although this discrepancy could be due to thiolation. The delivery of three different LNAs (Cur616, completely matching with the target, Cur222, presenting one mismatch, and Cur942, with four mismatches) for 14 days by an osmotic minipump in nude mice at different concentrations was followed by the analysis of the 15PC3 xenograft growth. Cur616 at 1 mg/kg clearly inhibited the tumour expansion and it was the most effective as compared to Cur222 and Cur942. The inhibition was dosedependent although increasing the dose caused a decrease in sequence specificity. Histological studies demonstrated that this treatment not only decreases the tumour size but also the number of viable tumour cells. With this work the authors demonstrated that LNAs are potent genotype-specific drugs that can be successfully utilised in cancer therapy[64]. |
| |
LNA SYNTHESIS
|
| |
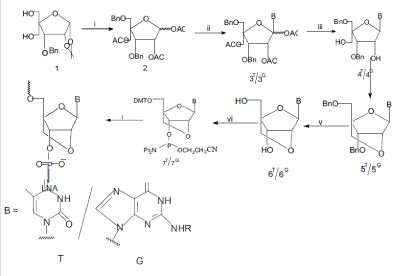
| Jesper Wengel et al reported the synthesis of a novel conformationally restricted nucleic acid mimic, LNA ( Scheme 1)[65,66,67]. Molecular modelling† and simple model building suggested to us that the LNA monomers would be favourably preorganized in an N-type conformation thus enabling the formation of entropically favoured duplexes with complementary DNA and RNA. As an attractive feature, the structural change from DNA (or RNA) to LNA is limited from a chemical perspective, namely the introduction of an additional 2A-C,4A-Coxymethylene link. |
| |
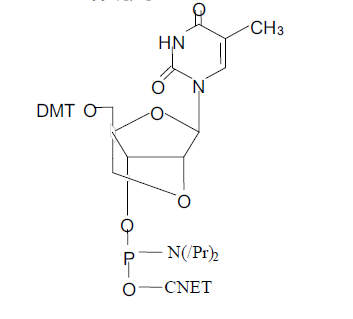
| Known 4A-C-hydroxymethyl pentofuranose derivative 1 was starting material for synthesis of LNA monomer (Scheme 1). Regioselective 5-Obenzylation, acetylation and acetolysis, followed by another acetylation, afforded furanose 2, a key intermediate for coupling with silylated nucleobases. Stereoselective reaction with silylated thymine10 yielded nucleoside 3T which was deacetylated to give nucleoside diol 4T. Tosylation followed by baseinduced ring closure afforded the 2A-O,4A-C-linked bicyclic nucleoside derivative 5T. Debenzylation yielded the unprotected analogue 6T as the first example of a nucleoside diol with the(1S,3R,4R,7S)- 7-hydroxy-1-hydroxymethyl-2,5-dioxabicyclo- [2.2.1]heptane structure. The assigned structure of 6T was verified by NMR spectroscopy.The absence of a coupling constant between 1A-H and 2A-H, and the unusual and strong mutual NOE effects (9%/8%) between 3A-H and 6-H (thymine base), strongly indicate structural preorganization of the pentofuranose ring of the LNA monomer into an Ntype conformation. Similar synthetic procedures were applied to synthesize the guanine derivatives 3G–6G via coupling of 2 and 2-Nisobutyrylguanine. Transformation of nucleosides 6 into the 5A-O-4,4A-dimethoxytrityl (5A-O-DMT) protected analogues and subsequently into the phosphoramidite derivatives 7§ yielded the desired monomeric building blocks for automated oligonucleotide synthesis. |
| |
 |
| |
| Scheme 1 Reagents and conditions (for 7T): |
| |
| i, (a) NaH, BnBr, DMF, |
| |
| (b) Ac2O, Py (64%, two steps), |
| |
| (c) 80% AcOH, |
| |
| (d) Ac2O, Py (86%, two steps); |
| |
| ii, thymine, N,O-bis(trimethylsilyl)acetamide, TMS triflate, acetonitrile |
| |
| (76%) [for 3G: 2-N-isobutyrylguanine, N,Obis (trimethylsilyl)acetamide, |
| |
| TMS triflate, dichloroethane]; |
| |
| iii, NaOMe, MeOH (97%); |
| |
| iv, (a) TsCl, Py (47%), |
| |
| (b) NaH, DMF (89%); |
| |
| v, 20% Pd(OH)2/C, EtOH, H2 (98%); |
| |
| vi (a) DMTCI, Py (93%), |
| |
| (b) N,N-diisopropylethylamine, 2-cyanoethyl N,N-diisopropylphosphoramidochloridite, CH2Cl2 (70%) |
| |
LNA THERMODYNAMICS
|
| |
| Singh et al (1998)[68] & Obika et al (1998)[69] reported on a minimal alteration of the pentose sugar of ribo- and deoxyribonucleotides that constrained, or “locked”, the sugar in the N-type confirmation seen in A-form DNA.The lock was achieved via a 2’- O,4’-C methylene linkage in 1,2:5,6-di-Oisopropylene- alpha-D-allofuranose. This alteration then served as the foundation for synthesizing locked nucleotide phosphoramidite monomers. Oligonucleotides containing one or more of these monomers were given the name locked nucleic acid ( Koshkin et al 1998)[70]. |
| |
 |
| |
| LNA were immediately seen to display remarkably increased thermodynamic stability and enhanced nucleic acid recognition.Initial investigstions of LNA, melting temperatures(Tm) by Koshkin et al.(1998)[70] revealed increased values per LNA monomer (Tm) of +3 to +5 C. and +4 to +8 C. against complementary DNA and RNA oligonucleotide respectively[75]. |
| |
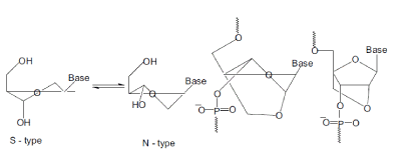
| Nucleic acid duplexes fall into two major conformational types, the A-type and the B-type, dictated by the puckering of the single nucleotides, a C3´-endo (N-type) conformation in the A-type and a C2´-endo (S-type) conformation in the B-type (Saenger, 1984)[71]. The A-type is adopted by RNA when dsRNA duplex regions are found, whereas the dsDNA in the genome adopt a B-type. Targeting DNA and RNA with high binding affinity can be effectuated by a conformational restriction in an Ntype conformation. |
| |
| In LNA, the O2´ and the C4´ atoms are linked by a methylene group hereby introducing a conformational lock of the molecule into a near perfect N-type conformation. LNA is oligonucleotides containing one or more of the 2´-O,4´-C-methylene- β-D-ribofuranosyl nucleosides called LNA monomers. |
| |
 |
| |
| Nucleoside Confor
mation & the Structure and the Locked Conformation of LNA Monomers
|
| |
| A major structural characteristic of LNA is its close resemblance to the natural nucleic acids. This leads to easy handling as LNA sequences have similar physical properties, including water solubility. Furthermore, LNA sequences are synthesised by the conventional phosphoramidite chemistry allowing automated synthesis of fully modified LNAsequences as well as chimeras with DNA, RNA, modified monomers or labels. |
| |
|
LNA Hybridisation
|
| |
| The most conclusive feature of LNA is the unprecedented hybridisation to complementary nucleic acids. In the following, some general observations on LNA hybridization are listed (Petersen and Wengel, 2003)[72]. |
| |
| • The increase in duplex stability by introducing one or several LNA monomers is strong in all cases with complementary RNA and in most cases with complementary DNA. |
| |
| • The affinity-enhancing effect pr. LNA monomer is generally more pronounced against complementary RNA than against complementary DNA (normally from +3 to +7 °C against RNA and from +2 to +5 °C against DNA). |
| |
| • LNA monomers incorporated in the 3´- or 5´- termini induce a smaller increase in thermal stability than do centrally placed LNA monomers. |
| |
| • The relative increase in thermal stability per LNA monomer is largest for a single or a few nonconsecutive incorporations in DNA/RNA/2’- OMe-RNA-sequences (mixmers). |
| |
| • The relative increase in thermal stability per LNA monomer is larger in shorter sequences than in longer sequences. |
| |
| • LNA monomers with pyrimidine bases tend to induce larger thermal stability increases than LNA monomers with purine bases (McTigue et al.,. 2004)[73]. |
| |
| • LNA monomers introduced into phosphorthioate sequences also strongly stabilise duplexes. |
| |
| • LNA monomers strongly stabilise triplexes and are useful in the design of triplex forming oligonucleotides (TFOs) composed of LNA mixmers with DNA. |
| |
| • LNA: LNA duplexes are inherently very stable, and to avoid application-limiting self-hybridizing this should be carefully considered when designing LNAsequences[ 74]. |
| |
CONCLUSION
|
| |
| LNA is a high afiinity , highly biostable DNA analogue. These features, combined with LNA’s close physical relatedness to DNA,make it highly attractive as a tool in biological research and DNA diagnostics and in the development of therapeutic drugs. It seems likely that many of the current technologies that uses DNA ,or other moderate affinity analogues, can be improved significantly by switching to LNA.It seems likely that LNA will facilitate the development of novel technologies that can not be achieved with standard DNA. |
| |
Conflict of Interest
|
| |
| NIL |
| |
Source of Support
|
| |
| NONE |
| |
| |
| |














