Keywords
Breast cancer; MCF-7; miRNA; Profiling; Biomarker
Introduction
Cancer describes a wide range of diseases that can affect different organs in the body. The normal behavior of cells is to divide and die in an orderly manner [1,2], however, cancer cells are capable to continually divide in an out-of-control way [3,4]. There are about 200 different types of cancers [5-7]. Cancer is the second leading cause of death globally with 8.8 million deaths every year [8].
Breast cancer, however, is the most common invasive malignant tumors and the second leading cause of death by cancer in females after lung cancer, with 1.5 million new cases per year and more than 500,000 cases globally per year [9-11]. It is a heterogeneous disease comprising various types of neoplasms, which involves different profile changes in both mRNA and micro-RNA (miRNA) expression [12-16].
To effectively monitor BC, early diagnosis with reliable tools and close observing of the patient’s response to treatment are needed [17,18]. However, these tasks are surprisingly difficult to tackle because of the lack of approved sensitive biomarkers to early detect or diagnose the disease [17,18]. A lot of evidence recently highlighted the potential use of miRNA in the early diagnosis of BC and other cancers [19-22]. miRNA also could be employed as a reliable biomarker, given the ease of isolation, characterization, and quantification. miRNA could not only be used in early diagnosis of BC but it also as prognostic or predictive biomarkers [22-24].
Extensive research in BC miRnome has yielded fascinating discoveries, some of which have as of late been approved to be utilized in clinical settings [23]. On-going miRNA research propels indicated incredible potential for the advancement of novel biomarkers and therapeutic targets [25-27].
miRNAs are a class of small single-stranded non-coding RNA molecules that are evolutionarily conserved and encoded by nearly 1% of the genome in most species. These are involved in the regulation of gene expression at the post-transcriptional level [28-30]. It is well known that some miRNAs function as key negative regulators in several biological processes underlie breast cancer [31]. Abnormal microRNA expression involved in the cancer process provides a suitable entry point to explore its functional role in cancer [13,32,33]. Deregulated miRNAs identified in BC might put us a step forward towards understanding the tumor microenvironment, which prompt investigate their role in cancer progression and spreading [34-36].
Several types of research have focused in the last decade to identify the association between different miRNA and BC via profiling the miRnome in either BC cells or patients [20,37-39]. This profiling helps in understanding the functional role and molecular mechanism of down/up-regulated microRNAs in BC development and progression, which, in turn, is needed to advance miRNA-based therapy [13,15,24].
Materials and Methods
Cell line maintenance
Breast cancer cell line (MCF-7) was obtained from the Holding Company for Biological Products and Vaccines (VACSERA, Giza, Egypt). Cells were seeded at a density of 104 cells/cm2 in 12-well plate and were cultured under standard laboratory conditions; at 37°C and 5% CO2 and maintained on RPMI 1640 media (GIBCO/Invitrogen Life Technologies, Carlsbad, USA) supplemented with 10% FBS (Hyclone, Logan, UT) and 1% antibiotic mix. MCF-7 cell line was enrolled in the study within the first 10 passages from the originally purchased flask to control genomic drift due to instability. RPMI-1640 was changed every 3 days and cells were passaged whenever it reached 65% to 80% confluently.
RNA extraction
Total RNA was extracted from the MCF-7 cells with the RNeasy kit (Qiagen, Germany). RNA was treated with DNase I (Boehringer-Mannheim, Mannheim, Germany) for 50 min. and purified according to the kit’s protocol. The quality and integrity of RNA were checked by spectrophotometry and ethidium bromide agarose gel electrophoresis.
First-strand cDNA synthesis
In a 0.2 mL PCR tube, the following items were combined: 200 ng of poly RNA, 0.6 L of 25 M semi-random primer, and ribonuclease-free water in a 4.75 L volume. The mix was incubated at 72°C in a hot-lid thermal cycler for 3 min., cooled down on the ice for 2 min., and incubated again at 25°C for 10 min. The following mixture was prepared and added to the tube: 2 L of 5x SMARTScribe buffer, 0.5L of 25 μM 5′SMART tag, 1 L of 10 mM deoxynucleotide triphosphate (dNTP) mix, 0.25 L of 100 mM DTT, 0.5 μL of RNaseOUT (Invitrogen), and 1 L of SMARTScribe Reverse Transcriptase (100 U) (Clontech). The mixture was incubated at 42°C for 90 min and then at 68°C for 10 min in a thermal block. To digest RNA, 1 L of RNase H (Invitrogen) was added to the mixture, and then the mixture was incubated at 37°C for an extra 20 min.
Double-stranded cDNA synthesis
About 11 μL of the prepared first-strand poly(A) cDNA was mixed with 74 μL of Milli-Q water, 10 μL of 10x PCR Buffer, 2 μL of 10 mM dNTP mix, 1 μL of 25 μM 5′ SMART PCR primer, and 2 μL of 50x Polymerase Mix (Clontech). A 100 μL volume of the reaction mixture for primer extension was incubated at 95°C for 1 min, 68°C for 20 min, and then 70°C for 10 min. Doublestranded cDNA was stored at -20°C until being used.
Real-time PCR
Using miScript PCR array for tumor suppressor genes (Qiagen, Germany), real-time-based expression analysis of 25 miRNA was performed in StepOne-Plus thermal cycler (Applied Biosystems). The thermal profile was as follows: 95°C for 1 min., 57°C for 45s, and 72°C for 1 min. The fold change was calculated using the 2- ΔCT method. U6 snRNA (as a housekeeping gene) was involved in the reaction to control the fidelity of the real-time amplification.
Data analysis
The retrieved data of CT values were uploaded to the online analysis tool provided by Qiagen [40], where the validation of data and calculation of 2-ΔΔCT for each miRNA were performed.
Statistical analysis
Statistical analyses were performed using the SPSS software package (SPSS, Inc., Chicago, IL). All values were expressed as mean ± SD. Analysis of variance with t-test was used to determine the significance of the difference in a multiple comparisons. Differences with a P value of less than 0.05 were considered statistically significant.
Target prediction
The up-regulated and down-regulated miRNAs in BC cells were submitted to miRNet.ca [41] online target prediction tool to identify the target gene(s) for both the up-and downregulated miRNAs.
Results
Up-regulated miRNA in BC
In the present study, 25 miRNAs were profiled in breast cancer cells (MCF-7). Eleven miRNA were found to be upregulated in BC cells; hsa-miR-141-3p, hsa-miR-16-5p, hsamiR- 196a-5p, hsa-miR-17-5p, hsa-miR-19a-3p, hsa-miR-181b-5p, hsa-miR-195-5p, hsa-miR-30b-5p, hsa-miR-103a-3p, hsamiR- 21-5p, and hsa-miR-30d-5p (Figure 1 and Table 1). Significant differences were obtained between U6 snRNA and all the miRNA studied (p=0.001), with one exception for hsa-141-3p where the differences was not significant at the same p-value.
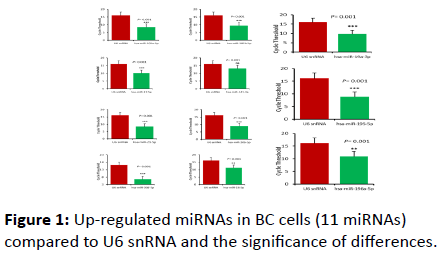
Figure 1: Up-regulated miRNAs in BC cells (11 miRNAs) compared to U6 snRNA and the significance of differences.
| miRNA name |
Mean |
p-value |
Sig. |
| hsa-let-7a-5p |
19.048 |
0.0848 |
ns |
| hsa-let-7b-5p |
37.972 |
0.0000 |
*** |
| hsa-miR-103a-3p |
8.583 |
0.0000 |
*** |
| hsa-miR-106a-5p |
30.777 |
0.0000 |
*** |
| hsa-miR-141-3p |
13.183 |
0.0723 |
ns |
| hsa-miR-143-3p |
35.751 |
0.0000 |
*** |
| hsa-miR-145-5p |
33.218 |
0.0000 |
*** |
| hsa-miR-17-5p |
10.083 |
0.0005 |
*** |
| hsa-miR-181b-5p |
9.484 |
0.0001 |
*** |
| hsa-miR-195-5p |
8.816 |
0.0000 |
*** |
| hsa-miR-196a-5p |
10.931 |
0.0023 |
** |
| hsa-miR-19a-3p |
9.742 |
0.0002 |
*** |
| hsa-miR-20b-5p |
37.413 |
0.0000 |
*** |
| hsa-miR-21-5p |
8.4139 |
0.0000 |
*** |
| hsa-miR-23b-3p |
30.629 |
0.0000 |
*** |
| hsa-miR-24-3p |
28.952 |
0.0000 |
*** |
| hsa-miR-30d-5p |
6.489 |
0.0000 |
*** |
| hsa-miR-31-5p |
28.750 |
0.0000 |
*** |
| hsa-miR-206 |
35.516 |
0.0000 |
*** |
| hsa-miR-30b-5p |
8.800 |
0.0000 |
*** |
| hsa-miR-99a-5p |
24.688 |
0.0000 |
*** |
| hsa-miR-133a |
18.307 |
0.1983 |
ns |
| hsa-miR-16-5p |
11.212 |
0.0037 |
** |
| hsa-miR-194-5p |
32.553 |
0.0000 |
*** |
| hsa-miR-30c-5p |
21.886 |
0.0010 |
*** |
| U6 snRNA |
16.179 |
-- |
-- |
Table 1: Statistical analysis of all miRNAs enrolled in this study (p=0.001).
Down regulated miRNAs in BC
Of the 25 miRNAs profiled In the present study, 14 miRNAs were found to be down-regulated in BC; hsa-miR-194-5p, hsamiR- 31-5p, hsa-miR-30c-5p, hsa-miR-99a-5p, hsa-miR-133a, hsamiR- 206, hsa-miR-23b-3p, hsa-miR-24-3p, hsa-miR-20b-5, hsamiR- 143-3p, hsa-miR-145-5p, hsa-miR-106a-5p, hsa-let-7a-5p, and hsa-let-7b-5p (Figure 2 and Table 1).
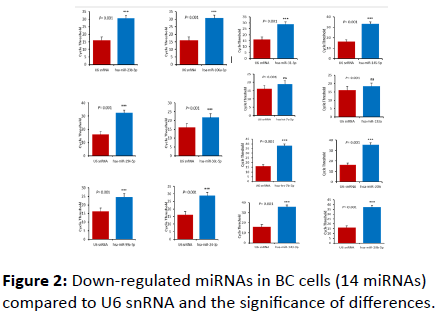
Figure 2: Down-regulated miRNAs in BC cells (14 miRNAs) compared to U6 snRNA and the significance of differences.
The differences between the level of expression of the target miRNA and U6 snRNA were significant (p=0.001). Two exceptions were recorded; hsa-let-7a-5p and hsa-133a where the differences in expression level were not significant at the same p-value (Figure 3).

Figure 3: Down-regulated and up-regulated miRNAs heat map based on CT values.
Target Prediction
The up- and down-regulated miRNAs detected in BC cells were uploaded to OmicsNet miRNA target prediction online tool (URL provided in the Materials and Methods section). Upregulated miRNAs were found to interact with several genes (Figure 4). FRS2, TNFRSF10B, BTBD7, MYC, and TPRG1L genes were the highlighted genes with which up-regulated miRNAs interact. The online database retrieved (for the 11 up-regulated miRNAs) 3202 nodes (miRNAs: 4, Targets: 3198) and 3718 edges.
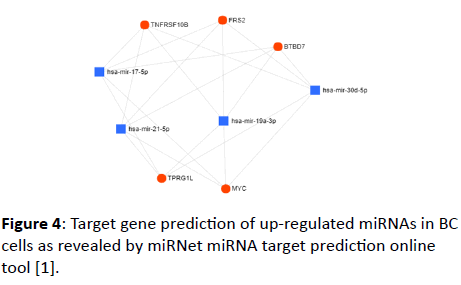
Figure 4: Target gene prediction of up-regulated miRNAs in BC cells as revealed by miRNet miRNA target prediction online tool [1].
For down-regulated miRNAs, a wide range of target genes was obtained (Figure 5). These target genes include GABPB2, GNB1, CCND1, ERCC1, ZNF417, GLO1, EIF4G2, DRAXIN, HOXD11, KPNA6, HSPA3, ZNF578, ACER2, CLS35F6, BDELK4, HIST1H2B, EIF4A2, COX6B1, HMGB1, HIF1A, ANKRD52, MFSD8, FOXK1, AGO3, CDKN1A, GATA6, and SLC30A7. The online database retrieved (for the 14 down-regulated miRNAs) 3079 nodes (miRNAs: 7, Targets: 3072) and 3608 edges. Two miRNAs with their target genes are shown in this study to avoid the complexity of the network retrieved using the seven miRNAs indented by the tool.
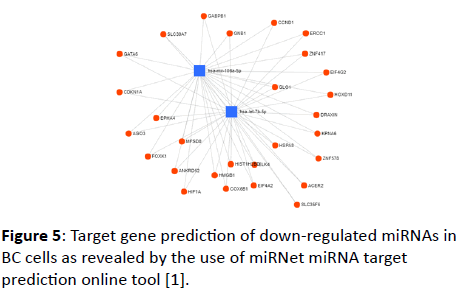
Figure 5: Target gene prediction of down-regulated miRNAs in BC cells as revealed by the use of miRNet miRNA target prediction online tool [1].
Disease correlation prediction
Besides predicting the target genes with which up- and downregulated miRNAs in BC cells interact, disease prediction also was performed using the same tool. hsa-miR-17-5p was presented as a model for the up-regulated miRNAs (Figure 6).
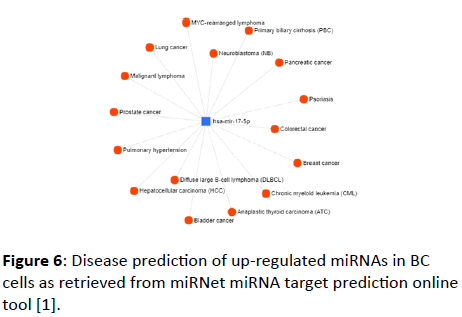
Figure 6: Disease prediction of up-regulated miRNAs in BC cells as retrieved from miRNet miRNA target prediction online tool [1].
A wide range of diseases was found to be correlated with the up-regulated miRNAs, these diseases include lymphoma, pancreatic cancer, lung cancer, prostate cancer, colorectal cancer, breast cancer, bladder cancer, and others.
Several diseases also were found to be correlated with the down-regulated miRNAs in BC cells (Figure 7); these diseases include leukemia, prostate cancer, hepatic cancer, ulcerative colitis, and others. hsa-miR-24-3p and hsa-miR-106a-5p were presented as representative of the down-regulated group of miRNAs.
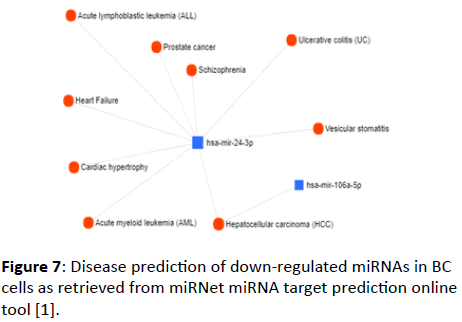
Figure 7: Disease prediction of down-regulated miRNAs in BC cells as retrieved from miRNet miRNA target prediction online tool [1].
Discussion
In the present study, 25 miRNAs were profiled in breast cancer cells (MCF-7) with the aim to identify a potential biomarker. Of these miRNAs, 14 were found to be downregulated and 11 were found to be up-regulated in BC cells as compared to U6 snRNA as an internal control in real time PCRbased expression analysis.
Up-regulated miRNAs
Statistical analysis revealed the significant differences between these miRNAs and U6 snRNA expect hsa-miR-141-3p where a non-significant difference was obtained (Table 1). This profile has been obtained earlier as Ranjha et al. [42] indicated that hsa-miR-141-3p was up-regulated in CRC, while Minn, Lee [43] indicated its elevated expression in breast cancer. hsamiR- 16-5p overexpression significantly inhibited the proliferation and colony formation in MCF-7 cells [44]. It has been indicated that apoptosis levels were significantly increased in hsa-miR-196a-5p mimic treated GC-2 cells, meaning that it is involved in the apoptotic machinery in cancer cells [45]. It could, likewise, be used as a potential cancer biomarker for digestive tract cancers [46]. The hsa-miR-17-5p and hsa-miR-21 were found to be elevated in breast cancer tissues and cell lines, where it down-regulates AIB1 gene primarily through translational inhibition. Meanwhile, hsa-miRNA-17-5p overexpression promotes cell proliferation and induces tumor growth [47-49]. The hsa-miR-30 was also found to play a role in neoplastic transformation, metastasis, and clinical outcomes in breast cancer development by controlling critical signaling pathways and relevant oncogenes. It also promotes the invasive phenotype of metastatic breast cancer cells by targeting NOV/ CCN3 [50].
Several up-regulated miRNAs i.e., hsa-miR-103a-3p hsamiR- 21-5p, and hsa-miR-195-5p could be employed as diagnostic biomarker for breast cancer [48,51-54].
The up-regulation of hsa-miRNA-19a-3p inhibited breast and bladder cancer progression and metastasis by inducing macrophage polarization through the down-regulated expression of Fra-1 proto-oncogene in vivo [55], and this miRNA might exhibit the same action in MCF-7 cells in vitro. It also plays a suppressive role in bone metastasis of prostate cancer, which makes it a potential candidate as an effective cancer therapy miRNA [56]. The oncogenic miR-181b was shown to be highly expressed in BC cells and many malignancies such as pancreatic, head and neck, and bladder cancer [57], as it mediates TGF-β1- induced epithelial-to-mesenchymal transition in non-small cell lung cancer stem-like cells derived from lung adenocarcinoma A549 cells, and this profile makes it a potential biomarker [58].
The hsa-miR-195-5p suppresses the proliferation, migration, and invasion of oral squamous cell carcinoma by targeting TRIM14 in vivo [59]. It works through the inhibition of tumor progression by targeting RPS6KB1 in cancer cells [60].
Down-regulated miRNAs
In the present investigation, of the 25 miRNAs studied, 14 were found to be down-regulated in BC cells. Although these 14 miRNAs were down-regulated in BC cells, some of them were up-regulated with tumor regulatory action such as hsa-miR-194 in gastric cancer [61].
hsa-mir-30c promotes the invasive phenotype of metastatic breast cancer cells by targeting NOV/CCN3 [48], meaning it must be up-regulated to perform its function, although it was downregulated in our study. Shukla et al. [62] also indicated that hsamiR- 30c negatively regulates NF-κB signaling and cell cycle progression through down-regulation of TRADD and CCNE1 in breast cancer. It has been indicated that hsa-miR-30 family is involved in the breast cancer development, and thus, it might serve as promising biomarkers and may bring a novel insight in molecular targeted therapy of breast cancer [63].
miR-99a has antitumor activity, and this activity is achieved by targeting the mTOR/p-4E-BP1/p-S6K1 pathway in human breast cancer cells [64], and by enhancing RAD001-induced apoptosis in human urinary bladder urothelial carcinoma cells [65]. These results were in accordance with the results obtained in the present study.
miR-133a was found to be up-regulated and negatively controls the expression of LASP1 in breast cancer cells [66], and it has potential use as a biomarker for breast cancer detection [67]. However, our investigation indicated an opposite profile for the expression of this microRNA. Nonetheless, our results were observed earlier when Wu et al. [68] indicated that miR-133a expression was associated with poor survival of breast cancer and restoration of miR-133a expression inhibited breast cancer cell growth and invasion [68].
Gao et al. [69] indicated that hsa-miR-206 represses the proliferation and invasion of breast cancer cells by targeting Cx43. This microRNA also inhibits the stemness and metastasis of breast cancer by targeting MKL1/IL11 pathway [70]. However, Zhou et al. [71] showed that it is down-regulated in breast cancer and found to inhibit cell proliferation through the upregulation of cyclinD2, and these results are in agreement with our results.
MicroRNA-23b is a tumor suppressor that functions by regulating Zeb1 in bladder cancer [72], cervical cancer [73], and ovarian cancer [74]. Behaving like a tumor suppressor miRNA indicates its down-regulation in cancer cells, and this profile was observed in the present study.
In cancer, loss-of-function of let-7 miRNAs has been linked to tumorigenesis via increased expression of target oncogenes [75]. MicroRNA let-7b regulates genomic balance by targeting Aurora B kinase [76]. Let-7, however, functions as a regulator of the ERα signaling pathway in human breast tumors and breast cancer stem cells [77], and acts as a novel suppressor by targeting the AKT2 gene [78]. Furthermore, let-7a suppresses breast cancer cell migration and invasion through down-regulation of C-C chemokine receptor type 7 [79]. These findings are in concordance with our obtained results in terms of the downregulation of let-7 miRNAs in BC cells.
Transcriptional profiling studies of miRNA expression across tumor tissues or cancer cell lines have revealed that miR-29 is down-regulated in the majority of cancers and up-regulated in the minority [80]. It regulates ER-positive breast cancer cell growth and invasion [81]. However, other study found that enforced miR-29a expression modulates apoptosis through inhibition of MCL-1 expression in ALCL cell lines, with a concomitant tumor growth reduction. Therefore, synthetic miR-29a might serve as a potential new tool for early diagnosis of cancer [82].
miR-24-3p functions as a tumor suppressor that is involved in cancer progression by regulating cell migration and invasion [83]. It might play a key role in breast cancer invasion and metastasis, and thus, could potentially be a target for cancer intervention [84]. miR-24 was down-regulated in BC cells where it may function as a tumor suppressor, in our study.
miR-24-3p acted as a tumor suppressor in hepatocellular carcinoma, as it inhibited cells proliferation through regulating CCDN1 and caspase3 expression [85]. It might function as a tumor suppressor in BC cells also as it found to be downregulated.
MicroRNA-20b was found to promote cell growth of breast cancer cells [86] and esophageal cancer cells [87] via targeting phosphatase and tensin homolog (PTEN). However, in our study, hsa-miR-20b was found to be down-regulated in BC cells.
hsa-miR-143 was markedly down-regulated in triple-negative breast cancer [88]. However, it inhibits cell proliferation and invasion by targeting DNMT3A in gastric cancer [89] with its tumor suppressor action [90], especially in BC cells [91].
microRNA-145 is Dual-strand tumor suppressor that functions via targeting MTDH in lung squamous cell carcinoma [92], and via inhibiting proliferation and migration of breast cancer cells by directly or indirectly regulating TGF-β1 expression [93].
Being capable to inhibit the proliferation and migration of astrocytoma cells [94], and invasion of renal cell carcinoma PAK5 [95], miR-106a-5p could be a potential tumor suppressor. This function is concordant with its low expression in BC as revealed in the present study.
Conclusion
In the present investigation, 25 miRNAs were profiled in MCF-7 breast cancer cells. Real-time PCR-based expression analysis data indicated that 11 miRNAs of the 25 studied miRNAs were up-regulated in BC cells, while 14 were down-regulated. Prediction analysis using miRNet online tool revealed that for up-regulated miRNAs were found to interact with several genes such as FRS2, TNFRSF10B, BTBD7, MYC, and TPRG1L. The downregulated miRNAs were found to interact with a wide array of genes that includes GABPB2, GNB1, CCND1, ERCC1, ZNF417, GLO1, EIF4G2, DRAXIN, HOXD11, KPNA6, HSPA3, ZNF578, ACER2, CLS35F6, BDELK4, HIST1H2B, EIF4A2, COX6B1, HMGB1, HIF1A, ANKRD52, MFSD8, FOXK1, AGO3, CDKN1A, GATA6, and SLC30A7. Based on disease prediction, the up-regulated miRNAs (hsa-miR-141-3p, hsa-miR-16-5p, hsa-miR-196a-5p, hsamiR- 17-5p, hsa-miR-19a-3p, hsa-miR-181b-5p, hsa-miR-195-5p, hsa-miR-30b-5p, hsa-miR-103a-3p, hsa-miR-21-5p, and hsamiR- 30d-5p) could be used as biomarkers in the early detection of BC. We could conclude that hsa-miR-30d-5p, the highly upregulated miRNA could essentially be employed as a potential diagnostic biomarker for BC. However, these data need further investigations to deeply identify the main role of this miRNA in developing/progression of breast cancer.
Acknowledgement
The authors thank Mahmoud Mohsen Mahmoud and Zeina Yasser for their help during the conduction of experiments.
Conflict of Interest
The authors declare no conflict of interests.
24417
References
- Fan Y, Siklenka K, Arora SK, Ribeiro P, Kimmins S, et al. (2016) miRNet-dissecting miRNA-target interactions and functional associations through network-based visual analysis. Nucleic Acids Res 44: 135-141.
- Pearson-Stuttard J, Zhou B, Kontis V, Bentham J, Gunter MJ, et al. (2018) Worldwide burden of cancer attributable to diabetes and high body-mass index: a comparative risk assessment. Lancet diabetes endocrinol 6: 6-15.
- Muller P, Walters S, Coleman MP, Woods L (2018) Which indicators of early cancer diagnosis from population-based data sources are associated with short-term mortality and survival? Cancer Epidemiology 56: 161-170.
- Ferlay J, Colombet M, Soerjomataram I, Dyba T, Randi G, et al. (2018) Cancer incidence and mortality patterns in Europe: estimates for 40 countries and 25 major cancers in 2018. Eur J Cancer 49: 1374-1403.
- Puente-Yagüe M, Cuadrado-Cenzual MA, Ciudad-Cabañas MJ, Hernández-Cabria M, Collado-Yurrita L (2018) Vitamin D: And its role in breast cancer. Kaohsiung J Med Sci 34: 423-427.
- Moses C, Garcia-Bloj B, Harvey AR, Blancafort P (2018) Hallmarks of cancer: The CRISPR generation. Eur J Cancer 93: 10-18.
- Siegel RL, Miller KD, Jemal A (2016) CA: A cancer journal for clinicians. Cancer stat 66: 7-30.
- Vasishta S, Ramesh S, Babu S, Ramakrishnegowda AS (2018) Awareness about breast cancer and outcome of teaching on breast self-examination in female degree college students. Indian J Med Res 9: 56-59.
- Taheripanah R, Balash F, Anbiaee R, Mahmoodi M, Sene AA (2018) Breast cancer and ovulation induction treatments. Clin breast cancer 18: 395-399.
- van Maaren MC, Lagendijk M, Tilanus-Linthorst MM, de Munck L, Pijnappel RM, et al. (2018) Breast cancer-related deaths according to grade in ductal carcinoma in situ: A Dutch population-based study on patients diagnosed between 1999 and 2012. Eur J Cancer 101: 134-142.
- Wu X, Zeng R, Wu S, Zhong J, Yang L, et al. (2015) Comprehensive expression analysis of miRNA in breast cancer at the miRNA and isomiR levels. Gene 557: 195-200.
- Wang M, Cai WR, Meng R, Chi JR, Li YR, et al. (2018) miR-485-5p suppresses breast cancer progression and chemosensitivity by targeting survivin. Biochem biophys res commun 501: 48-54.
- Di Leva G, Croce CM (2010) Roles of small RNAs in tumor formation. Trends Mol Med 16: 257-267.
- Rohanizadegan M (2018) Analysis of circulating tumor DNA in breast cancer as a diagnostic and prognostic biomarker. Cancer Genet 228: 159-168.
- Sharma B, Kanwar SS (2018) Phosphatidylserine: A cancer cell targeting biomarker. Semin cancer biol 52: 17-25.
- Chin AR, Fong MY, Somlo G, Wu J, Swiderski P, et al. (2016) Cross-kingdom inhibition of breast cancer growth by plant miR159. Cell Res 26: 217.
- Elghoroury EA, ElDine HG, Kamel SA, Abdelrahman AH, Mohammed A, et al. (2018) Evaluation of miRNA-21 and miRNA Let-7 as prognostic markers in patients with breast cancer. Clin Breast Cancer 18: 721-726.
- Zhang B, Pan X, Cobb GP, Anderson TA (2007) microRNAs as oncogenes and tumor suppressors. Dev Biol 302: 1-2.
- Sherafatian M (2018) Tree-based machine learning algorithms identified minimal set of miRNA biomarkers for breast cancer diagnosis and molecular subtyping. Gene 677: 111-118.
- Ivanov YD, Pleshakova TO, Malsagova KA, Kozlov AF, Kaysheva AL, et al. (2018) Detection of marker miRNAs in plasma using SOI-NW biosensor. Sens Actuators B Chem 261: 566-571.
- Blenkiron C, Miska EA (2007) miRNAs in cancer: approaches, aetiology, diagnostics and therapy. Hum Mol Genet 16: 106-113.
- Abba ML, Patil N, Leupold JH, Moniuszko M, Utikal J, et al. (2017) MicroRNAs as novel targets and tools in cancer therapy. Cancer Lett 387: 84-94.
- Garofalo M, Croce CM (2013) MicroRNAs as therapeutic targets in chemoresistance. Drug Resist Updat 16: 47-59.
- https://www.elsevier.com/books/comprehensive-biomaterials-ii/ducheyne/978-0-08-100691-7
- Velmurugan G, Dasgupta A, Krishnan K, Sivakumar A, Yacob JC, et al. (2017) Functional Genomics of MicroRNAs. Curr Develop Biotechnol Bioeng 103-121.
- https://www.elsevier.com/books/rna-methodologies/farrell-jr/978-0-12-804678-4.
- Mekala JR, Naushad SM, Ponnusamy L, Arivazhagan G, Sakthiprasad V, et al. (2018) Epigenetic regulation of miR-200 as the potential strategy for the therapy against triple-negative breast cancer. Gene 641: 248-258.
- Fabbri M, Calin GA (2010) Epigenetics and miRNAs in human cancer. Adv Genet 70: 87-99.
- Long H, Wang X, Chen Y, Wang L, Zhao M, et al. (2018) Dysregulation of microRNAs in autoimmune diseases: pathogenesis, biomarkers and potential therapeutic targets. Cancer Lett 428: 90-103.
- Kai K, Dittmar RL, Sen S (2018) Secretory microRNAs as biomarkers of cancer. Semin Cell Dev Biol 78: 22-36.
- Gao Y, Lin L, Li T, Yang J, Wei Y (2017) The role of miRNA-223 in cancer: Function, diagnosis and therapy. Gene 616: 1-7.
- Loginov VI, Pronina IV, Burdennyy AM, Filippova EA, Kazubskaya TP, et al. (2018) Novel miRNA genes deregulated by aberrant methylation in ovarian carcinoma are involved in metastasis. Gene 662: 28-36.
- Pronina IV, Loginov VI, Burdennyy AM, Fridman MV, Senchenko VN, et al. (2017) DNA methylation contributes to deregulation of 12 cancer-associated microRNAs and breast cancer progression. Gene 604: 1-8.
- Chen J, Jiang Y, Zhou J, Liu S, Qin N, et al. (2018) Evaluation of CpG-SNPs in miRNA promoters and risk of breast cancer. Gene 651: 1-8.
- Katar S, Baran O, Evran S, Cevik S, Akkaya E, et al. (2017) Expression of miRNA-21, miRNA-107, miRNA-137 and miRNA-29b in meningioma. Clin Neurol Neurosurg 156: 66-70.
- https://www.qiagen.com/eg/shop/genes-and-pathways/data-analysis-center-overview-page/miscript-mirna-pcr-array-data-analysis-center
- Ranjha R, Aggarwal S, Bopanna S, Ahuja V, Paul J (2015) Site-specific microRNA expression may lead to different subtypes in ulcerative colitis. PloS One 10: e0142869.
- Minn YK, Lee DH, Hyung WJ, Kim JE, Choi J, et al. (2014) MicroRNA-200 family members and ZEB2 are associated with brain metastasis in gastric adenocarcinoma. Int J Oncol 45: 2403-2410.
- Qu Y, Liu H, Lv X, Liu Y, Wang X, et al. (2017) MicroRNA-16-5p overexpression suppresses proliferation and invasion as well as triggers apoptosis by targeting VEGFA expression in breast carcinoma. Oncotarget 8: 72400.
- Lu J, Gu H, Tang Q, Wu W, Yuan B, et al. (2016) Common SNP in hsa-miR-196a-2 increases hsa-miR-196a-5p expression and predisposes to idiopathic male infertility in Chinese Han population. Sci Rep 6: 19825.
- Lu YC, Chang JT, Chan EC, Chao YK, Yeh TS, et al. (2016) miR-196, an emerging cancer biomarker for digestive tract cancers. J Cancer 7: 650.
- Hossain A, Kuo MT, Saunders GF (2006) Mir-17-5p regulates breast cancer cell proliferation by inhibiting translation of AIB1 mRNA. Mol Cell Biol 26: 8191-8201.
- Yang F, Li Y, Xu L, Zhu Y, Gao H, et al.( 2017) miR-17 as a diagnostic biomarker regulates cell proliferation in breast cancer. Onco Targets Ther 10: 543.
- Yanwirasti WA, Arisanty D (2017) Evaluation of MiR-21 and MiR-10b expression of human breast cancer in west Sumatera. Pak J Biol Sci 20: 189-196.
- Dobson JR, Taipaleenmäki H, Hu YJ, Hong D, Van Wijnen AJ, et al. (2014) hsa-mir-30c promotes the invasive phenotype of metastatic breast cancer cells by targeting NOV/CCN3. Cancer Cell Int 14: 73.
- Weber DG, Casjens S, Johnen G, Bryk O, Raiko I, et al. (2014) Combination of MiR-103a-3p and mesothelin improves the biomarker performance of malignant mesothelioma diagnosis. PloS One 9: e114483.
- Zheng R, Liu Y, Zhang X, Zhao P, Deng Q (2017) miRNA-200c enhances radiosensitivity of esophageal cancer by cell cycle arrest and targeting P21. Biomed Pharmacother 90: 517-523.
- Tanjung DS, Fitria MS, Kartika AI, Rakhmina D, Anwar SL, et al. (2014) The expression of hsa-mir-21-5p as minimal invasive marker to adjuvant chemotherapy in breast cancer patients. J Med Sci 48.
- Luo Q, Wei C, Li X, Li J, Chen L, et al. (2014) MicroRNA-195-5p is a potential diagnostic and therapeutic target for breast cancer. Oncol Rep 31: 1096-1102.
- Feng Y, Liu J, Kang Y, He Y, Liang B, et al. (2014) miR-19a acts as an oncogenic microRNA and is up-regulated in bladder cancer. J Exp Clin Cancer Res 33: 67.
- Huang S, Wa Q, Pan J, Peng X, Ren D, et al. (2017) Downregulation of miR-141-3p promotes bone metastasis via activating NF-κB signaling in prostate cancer. J Exp Clin Cancer Res 36: 173.
- Liu J, Shi W, Wu C, Ju J, Jiang J (2014) miR‑181b as a key regulator of the oncogenic process and its clinical implications in cancer. Biomed Rep 2: 7-11.
- Li X, Han J, Zhu H, Peng L, Chen Z (2017) miR‑181b‑5p mediates TGF‑β1-induced epithelial-to-mesenchymal transition in non-small cell lung cancer stem-like cells derived from lung adenocarcinoma A549 cells. Int J Oncol 51: 158-168.
- Wang T, Ren Y, Liu R, Ma J, Shi Y, et al. (2017) miR-195-5p suppresses the proliferation, migration, and invasion of oral squamous cell carcinoma by targeting TRIM14. Biomed Res Int 1-13.
- Cai C, Chen QB, Han ZD, Zhang YQ, He HC, et al. (2015) miR-195 inhibits tumor progression by targeting RPS6KB1 in human prostate cancer. Clin Cancer Res 21: 4922-4934.
- Yang R, Yang F, Huang Z, Jin Y, Sheng Y, et al. (2017) Serum microRNA-122-3p, microRNA-194-5p and microRNA-5099 are potential toxicological biomarkers for the hepatotoxicity induced by Airpotato yam. Toxicol Lett 280: 125-132.
- Shukla K, Sharma AK, Ward A, Will R, Hielscher T, et al. (2015) MicroRNA‐30c‐2‐3p negatively regulates NF‐κB signaling and cell cycle progression through downregulation of TRADD and CCNE1 in breast cancer. Mol Oncol 9: 1106-1119.
- Yang SJ, Yang SY, Wang DD, Chen X, Shen HY, et al. (2017) The miR-30 family: Versatile players in breast cancer. Tumour Biol 39: 1-13.
- Hu Y, Zhu Q, Tang L (2014) MiR-99a antitumor activity in human breast cancer cells through targeting of mTOR expression. PloS One 9: 1-10.
- Tsai TF, Lin JF, Chou KY, Lin YC, Chen HE, et al. (2018) miR-99a-5p acts as tumor suppressor via targeting to mTOR and enhances RAD001-induced apoptosis in human urinary bladder urothelial carcinoma cells. Onco Targets Ther 11: 239.
- Sui Y, Zhang X, Yang H, Wei W, Wang M (2018) MicroRNA-133a acts as a tumour suppressor in breast cancer through targeting LASP1. Oncology Report 39: 473-482.
- Shen J, Hu Q, Schrauder M, Yan L, Wang D, et al. (2014) Circulating miR-148b and miR-133a as biomarkers for breast cancer detection. Oncotarget 5: 5284.
- Wu ZS, Wang CQ, Xiang R, Liu X, Ye S, et al. (2012) Loss of miR-133a expression associated with poor survival of breast cancer and restoration of miR-133a expression inhibited breast cancer cell growth and invasion. BMC cancer 12: 51.
- Gao P, He M, Zhang C, Geng C. (2018) Integrated analysis of gene expression signatures associated with colon cancer from three datasets. Gene 654: 95-102.
- Samaeekia R, Adorno-Cruz V, Bockhorn J, Chang YF, Huang S, et al. (2017) miR-206 inhibits stemness and metastasis of breast cancer by targeting MKL1/IL11 pathway. Clin Cancer Res 3: 1091-1103.
- Zhou J, Tian Y, Li J, Lu B, Sun M, et al. (2013) miR-206 is down-regulated in breast cancer and inhibits cell proliferation through the up-regulation of cyclinD2. Biochem Biophys Res Commun 433: 207-212.
- Majid S, Dar AA, Saini S, Deng G, Chang I, et al. (2013) MicroRNA-23b functions as a tumor suppressor by regulating Zeb1 in bladder cancer. PloS One 8: 1-9.
- Campos-Viguri GE, Jiménez-Wences H, Peralta-Zaragoza O, Torres-Altamirano G, Soto-Flores DG, et al. (2015) miR-23b as a potential tumor suppressor and its regulation by DNA methylation in cervical cancer. Infect Agent Cancer 10: 42.
- Li W, Liu Z, Chen L, Zhou L, Yao Y (2014) MicroRNA-23b is an independent prognostic marker and suppresses ovarian cancer progression by targeting runt-related transcription factor-2. FEBS Letter 588: 1608-1615.
- Qattan A, Intabli H, Alkhayal W, Eltabache C, Tweigieri T, et al. (2017) Robust expression of tumor suppressor miRNA’s let-7 and miR-195 detected in plasma of Saudi female breast cancer patients. BMC Cancer 17: 799.
- Mäki-Jouppila JH, Pruikkonen S, Tambe MB, Aure MR, Halonen T, et al. (2015) MicroRNA let-7b regulates genomic balance by targeting Aurora B kinase. Mol Oncol 9: 1056-1070.
- Sun X, Qin S, Fan C, Xu C, Du N, et al. (2013) Let-7: a regulator of the ERα signaling pathway in human breast tumors and breast cancer stem cells. Oncology Report 29: 2079-2087.
- Zhou B, Shan H, Su Y, Xia K, Zou R, et al. (2017) Let-7a inhibits migration, invasion and tumor growth by targeting AKT2 in papillary thyroid carcinoma. Oncotarget 8: 1-10.
- Kim SJ, Shin JY, Lee KD, Bae YK, Sung KW, et al. (2012) MicroRNA let-7a suppresses breast cancer cell migration and invasion through downregulation of CC chemokine receptor type 7. Breast Cancer Res 14: R14.
- Jiang H, Zhang G, Wu JH, Jiang CP (2014) Diverse roles of miR-29 in cancer. Oncology Report 31: 1509-1516.
- Li ZH, Xiong QY, Xu L, Duan P, Yang QO, et al. (2017) miR-29a regulated ER-positive breast cancer cell growth and invasion and is involved in the insulin signaling pathway. Oncotarget 8: 32566.
- Desjobert C, Renalier MH, Bergalet J, Dejean E, Joseph N, et al. (2011) MiR-29a down-regulation in ALK-positive anaplastic large cell lymphomas contributes to apoptosis blockade through MCL-1 overexpression. Blood 117: 6627-6637.
- Kang H, Rho JG, Kim C, Tak H, Lee H, et al. (2017) The miR-24-3p/p130Cas: a novel axis regulating the migration and invasion of cancer cells. Sci Rep 7: 1-10.
- Du WW, Fang L, Li M, Yang X, Liang Y, et al. (2013) MicroRNA miR-24 enhances tumor invasion and metastasis by targeting PTPN9 and PTPRF to promote EGF signaling. J Cell Sci 126: 1440-1453.
- Khodadadi-Jamayran A, Akgol-Oksuz B, Afanasyeva Y, Heguy A, Thompson M, et al. (2018) Prognostic role of elevated mir-24-3p in breast cancer and its association with the metastatic process. Oncotarget 9: 12868.
- Zhou W, Shi G, Zhang Q, Wu Q, Li B, et al. (2014) MicroRNA-20b promotes cell growth of breast cancer cells partly via targeting phosphatase and tensin homologue (PTEN). Cell Biosci 4: 62.
- Wang B, Yang J, Xiao B (2016) MicroRNA-20b (miR-20b) promotes the proliferation, migration, invasion, and tumorigenicity in esophageal cancer cells via the regulation of phosphatase and tensin homologue expression. PloS One 11: 1-19.
- Chang YY, Kuo WH, Hung JH, Lee CY, Lee YH, et al. (2015) Deregulated microRNAs in triple-negative breast cancer revealed by deep sequencing. Mol Cancer 14: 36.
- Zhang Q, Feng Y, Liu P, Yang J (2017) MiR-143 inhibits cell proliferation and invasion by targeting DNMT3A in gastric cancer. Tumour Biol 39: 1-8.
- Johannessen C, Moi L, Kiselev Y, Pedersen MI, Dalen SM, et al. (2017) Expression and function of the miR-143/145 cluster in vitro and in vivo in human breast cancer. PloS One 12: 1-22.
- He Z, Yi J, Liu X, Chen J, Han S, et al. (2016) MiR-143-3p functions as a tumor suppressor by regulating cell proliferation, invasion and epithelial-mesenchymal transition by targeting QKI-5 in esophageal squamous cell carcinoma. Mol Cancer 15: 51.
- Mataki H, Seki N, Mizuno K, Nohata N, Kamikawaji K, et al. (2016) Dual-strand tumor-suppressor microRNA-145 (miR-145-5p and miR-145-3p) coordinately targeted MTDH in lung squamous cell carcinoma. Oncotarget 7: 72084.
- Ding Y, Zhang C, Zhang J, Zhang N, Li T, et al. (2017) miR-145 inhibits proliferation and migration of breast cancer cells by directly or indirectly regulating TGF-β1 expression. Int J Oncol 50: 1701-1710.
- Zhi F, Zhou G, Shao N, Xia X, Shi Y, et al. (2013) miR-106a-5p inhibits the proliferation and migration of astrocytoma cells and promotes apoptosis by targeting FASTK. PLoS One 8: 1-9.
- Pan YJ, Wei LL, Wu XJ, Huo FC, Mou J, et al. (2017) MiR-106a-5p inhibits the cell migration and invasion of renal cell carcinoma through targeting PAK5. Cell Death Dis 8: 1-12.