Birgit Voigt1*, Dirk Albrecht1 and Axel Dalhoff2
1Institute of Microbiology, Ernst-Moritz-Arndt University, Greifswald, Germany
2Institute for Infection Medicine, Christian-Albrechts University, Kiel, Germany
*Corresponding Author:
Birgit Voigt, PhD
Ernst-Moritz-Arndt University
Institute for Microbiology
F.-L.-Jahn-Str. 15
D-17487 Greifswald, Germany
Tel: +49-3834-864237
FAX: +49-3834-864202
E-mail: birgit.voigt@uni-greifswald.de
Received date: November 10, 2016; Accepted date: December 15, 2016; Published date: December 22, 2016
Citation: Voigt B, Albrecht D, Dalhoff A. Mode of Action of MCB3681 in Staphylococcus aureus–A Proteomic Study. Arch Clin Microbiol. 2016, 7:6.
Copyright: © 2016 Voigt B, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Keywords
Quinolone; Staphylococcus aureus; 2D-Gel electrophoresis; Protein expression; Proteome
Introduction
Proteomics is a powerful tool to study differential protein expression of bacteria in response to antibiotics. Studying changes of the bacterial proteome in response to antibiotics allows conclusions about the mode of action and the targets of the antibiotic [1-5].
MCB3681 is a novel small molecule with structural elements of an oxazolidinone and a quinolone, showing good activity against Gram-positive bacteria irrespective of whether the isolates are quinolone- and/or linezolid-resistant [6,7]. The minimal inhibition concentrations (MICs) of MCB3681 are significantly lower than those of the 1:1 combination of ciprofloxacin and linezolid, thus demonstrating that its intrinsic activity is qualitatively and quantitatively different from oxazolidinones and quinolones. From MCB3681’s activity against ciprofloxacin and linezolid double-resistant mutants it may be hypothesized, that MCB3681 interacts with targets other than the known quinolone and oxazolidinone targets [6,7]. Therefore, the aim of this study was to examine if MCB3681 may affect expression of proteins other than those affected by ciprofloxacin or linezolid.
Methods
The quality-control strain S. aureus ATCC 29213 and the test strain S. aureus RN1HG001 were grown aerobically under vigorous agitation at 37°C in a defined medium [8,9]. Strain RN1HG001 was used because it is susceptible to most antibiotics. In strain RN1HG001 the rsbU gene of S. aureus RN1 (NCTC8325), a strain widely used in research projects, is repaired [9]. MICs were determined in duplicate according to guidelines of the Clinical and Laboratory Standards Institute (CLSI) [10].
Proteins were pulse labeled with L-[35S]-methionine according to Engelmann et al. [11]. Cells at an OD500 0.4 were exposed to either 4 or 20 mg/L MCB3681 for 5 min before adding labeled methionine (3.7 MBq). Incorporation was stopped 5 min later by adding excess unlabeled methionine and chloramphenicol. Preparation of cytosolic protein extracts, protein quantification and measurement of methionine incorporation were done according to Reiss et al. [12], except that for disruption of bacteria a Precellys 24 homogenizer (Peqlab) was used twice for 30 s at 6800 rpm, separated by 5 min incubation on ice. Proteins for preparative 2D gels were obtained from bacteria exposed at an OD500 0.4 to 4 mg/L MCB3681 for 30 min.
Protein extracts (100 μg for radiolabeled samples, 300 μg for preparative gels) were separated using commercially available IPG-strips in the pH range of 4-7 [13]. Labeled gels were dried on a heated vacuum dryer followed by exposure to storage phosphor screens. Screens were scanned using a Typhoon Scanner according to Reiss et al. [12]. Preparative gels were stained and scanned as described previously [14]. For protein identification, spots were cut from preparative gels. Mass spectrometry was performed according to Wolf et al. [15]. A database including complete or partial sequences of more than 100 S. aureus strains available from the NCBI website was used for searching. Search parameters were the same as described in previous studies [15]. Only proteins whose synthesis was induced or repressed at least twice in all three biological replicates were identified. Gels were analyzed using the Delta2D software (Decodon) [16].
Results and Discussion
Both S. aureus strains are inhibited by 0.25 mg/L MCB3681 and concentrations up to 10 mg/L are bacteriostatic while an immediate inhibition of protein synthesis was observed at 20 mg/L (Figure S1). Stained gels of protein samples exposed to 20 mg/L MCB3681 resemble the controls. One structural element of MCB3681 is part of an oxazolidinone which are known to inhibit protein synthesis almost immediately, with the effect that the proteomes are “frozen in time” [17,18]. Therefore, labeling experiments were repeated with 4 mg/L MCB3681. Synthesis of most proteins is not affected by MCB3681 (Figure 1). Synthesis of 13 proteins is induced, and synthesis of 16 proteins is repressed. A total of 27 out of 29 protein spots with altered synthesis could be identified; all are cytosolic proteins (Table 1 and Table S1).
| Label |
Annotated function |
Ratio |
Ratio |
Ratio |
Mean |
Standard deviation |
| control1_ |
control1_ |
control1_ |
| vs_MCB 1 |
vs_MCB 2 |
vs_MCB 3 |
| Induced synthesis of proteins |
| RpsF1 |
30S ribosomal protein S6 |
4.8 |
4.85 |
4.84 |
4.83 |
0.02 |
| SucD |
succinyl-CoA ligase [ADP-forming] subunit alpha |
3.97 |
2.38 |
3.04 |
3.13 |
0.65 |
| RpsF |
30S ribosomal protein S6 |
3.85 |
4.53 |
3.95 |
4.11 |
0.3 |
| RplJ |
50S ribosomal protein L10 |
3.64 |
3.71 |
3.14 |
3.5 |
0.25 |
| PyrR |
pyrimidine operon regulatory protein |
3.51 |
2.63 |
2.8 |
2.98 |
0.38 |
| FusA |
elongation factor G |
3.49 |
3.09 |
4.06 |
3.55 |
0.4 |
| FabF1 |
3-oxoacyl-[acyl-carrier-protein] synthase 2 |
2.77 |
2.66 |
2.45 |
2.62 |
0.13 |
| RplL |
50S ribosomal protein L7/L12 |
2.71 |
3.03 |
2.82 |
2.85 |
0.13 |
| SAR1041/ PurQ |
hypothetical/ phospho- ribosylformylglycinamidine synthase 1 |
2.34 |
2.18 |
2.23 |
2.25 |
0.07 |
| SACOL2532 |
acetyltransferase |
2.19 |
2.39 |
2.26 |
2.28 |
0.08 |
| SACOL2379 |
hypothetical |
2.09 |
2.31 |
2.25 |
2.22 |
0.09 |
| FtnA |
ferritin |
2.08 |
2.9 |
2.76 |
2.58 |
0.36 |
| Protein synthesis not affected |
| FabF |
3-oxoacyl-[acyl-carrier-protein] synthase 2 |
1.23 |
1.39 |
1.46 |
1.36 |
0.1 |
| Tuf |
elongation factor Tu |
1.05 |
1.15 |
1.24 |
1.15 |
0.08 |
| Reduced synthesis of proteins |
| SACOL0426 |
acetyl-CoA acetyltransferase |
0.41 |
0.45 |
0.48 |
0.44 |
0.03 |
| KatA |
catalase |
0.38 |
0.3 |
0.36 |
0.35 |
0.04 |
| Pyk |
pyruvate kinase |
0.39 |
0.41 |
0.44 |
0.42 |
0.02 |
| PepF |
oligoendopeptidase F |
0.39 |
0.42 |
0.44 |
0.42 |
0.02 |
| MetE1 |
5-methyltetrahydropteroyl- triglutamate-homocysteine methyltransferase |
0.38 |
0.3 |
0.51 |
0.4 |
0.09 |
| GlyA |
serine hydroxymethyltransferase |
0.33 |
0.09 |
0.46 |
0.29 |
0.15 |
| GcvPB |
glycine dehydrogenase [decarboxylating] subunit 2 |
0.32 |
0.37 |
0.58 |
0.42 |
0.11 |
| IleS |
isoleucine-tRNA ligase |
0.32 |
0.24 |
0.35 |
0.3 |
0.05 |
| GlnA |
glutamine synthetase |
0.32 |
0.27 |
0.45 |
0.35 |
0.08 |
| Pyc |
pyruvate carboxylase |
0.29 |
0.3 |
0.62 |
0.4 |
0.15 |
| AspS |
aspartate-tRNA ligase |
0.28 |
0.42 |
0.43 |
0.38 |
0.07 |
| IleS1 |
isoleucine-tRNA ligase |
0.24 |
0.23 |
0.19 |
0.22 |
0.02 |
| RpsA |
30S ribosomal protein S1 |
0.24 |
0.28 |
0.46 |
0.33 |
0.09 |
| MetE |
5-methyltetrahydropteroyl- triglutamate-homocysteine methyltransferase |
0.22 |
0.23 |
0.23 |
0.23 |
0 |
| FemB |
Aminoacyl transferase |
0.09 |
0.33 |
0.42 |
0.28 |
0.14 |
Table 1 MCB3681/control synthesis ratios of proteins in MCB3681 treated S. aureus N1HG001 cells (ratios of ≤ 0.5 represent a reduced synthesis of proteins, ratios of >2 represent an induced synthesis of proteins).
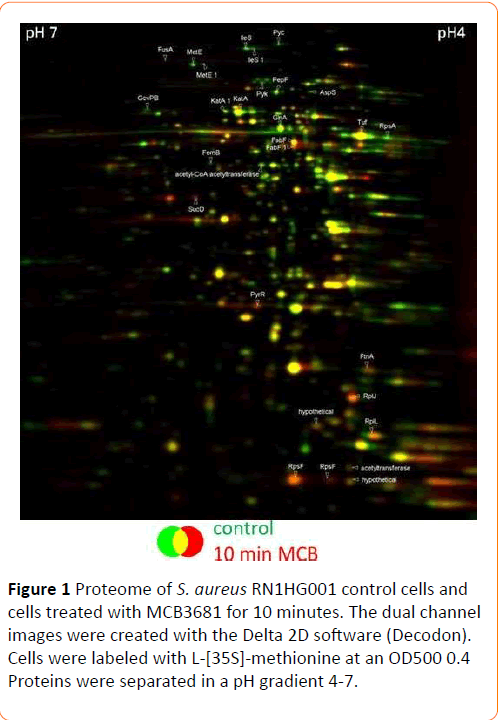
Figure 1: Proteome of S. aureus RN1HG001 control cells and cells treated with MCB3681 for 10 minutes. The dual channel images were created with the Delta 2D software (Decodon). Cells were labeled with L-[35S]-methionine at an OD500 0.4 Proteins were separated in a pH gradient 4-7.
Among the induced proteins are the bifunctional protein PyrR and the ferritin family protein FtnA. FabF, 3-oxoacyl-[acyl-carrierprotein] synthase 2, occurrs in two spots, only one of these spots is induced. Synthesis of four ribosomal proteins (RplL, RpsF, RplJ, RpsF) is induced, too. One ribosomal protein (RpsA) and two aminoacyl-tRNA synthetases (IleS, AspS) are synthesized at a lower level. Synthesis of ribosomal proteins is induced immediately while synthesis of most aminoacyl-tRNAsynthetases is reduced as a downstream-effect by translation inhibitors like linezolid [19] or amino-acid starvation. However, MCB3681 reduces synthesis of the isoleucyl-tRNA-synthetase and aspartyl-tRNA-synthetase instantly. This strongly reduced expression of two aminoacyl-tRNA-synthetases may represent an additional mode of action of MCB3681.
Synthesis of proteins from different amino acid synthesis pathways (GlyA, MetE, GlnA) is reduced in MCB3681 treated cells. Furthermore, exposure to MCB3681 causes a significantly reduced synthesis of methicillin-resistance factor protein FemB and of catalase, KatA, which forms three distinct spots in the gel. The reduced expression of the femB and katA gene products may be clinically relevant. Methicillin resistance factor proteins A and B are equally important for expression of methicillinresistance [20]. Reduced expression of either of these proteins results in an increased β-lactam susceptibility [20]. Reduced synthesis of FemB due to exposure to MCB3681 likely contributes to its pronounced activity against MRSA. KatA on the other hand is required for nasal colonization [17]. Thus, MCB3681 may modulate adhesion of pathogens to mucosal surfaces.
Analysis of protein expression patterns does not reveal any effects of MCB3681 on DNA-replication mechanisms and the SOS-response although one structural element of MCB3681 is part of a quinolone. These findings are, nevertheless, in agreement with its mode of action. The biochemical analysis revealed that affinity of MCB3681 to topoisomerase IV is low and affinity to DNA-gyrase is 10-times lower than that of ciprofloxacin [18]. Transcriptional profiling of ciprofloxacin’s effect on gene-expression revealed that only 20% of the genes involved in translation were repressed [1].
Both, the moderate effect of ciprofloxacin on the translational apparatus and the moderate affinity of MCB3681 to type II topoisomerases may explain why an effect of MCB3681 on the DNA replication machinery is not noted in this study. Likewise, the SOS-response is induced moderately by sub-inhibitory concentrations of MCB3681 whereas supra-inhibitory MCB3681 concentrations have no effect on the SOS-response [18]. The indicator strain used in this study has been exposed to suprainhibitory MCB3681 concentrations for 10 minutes only, thus avoiding falsifying secondary effects resulting from downstream effects triggered by target inhibition. Therefore, short-term exposure to MCB3681 does not affect SOS-response.
Conclusions
In summary, addition of MCB3681 to growing S. aureus cells causes a moderate reprogramming of protein synthesis with 29 protein spots exhibiting altered synthesis. In contrast, linezolid [19] and ciprofloxacin [21] provoked a much higher number of changes in gene expression. Following linezolid- and ciprofloxacin-stress, expression of 566 [19] and 235 proteins [21], respectively, is changed. Thus, the impact of MCB3681 on the proteome signature is different from that of either ciprofloxacin or linezolid.
Acknowledgement
We thank Susanne Gebauer for excellent technical assistance.
Funding
This work was funded by the Morphochem Aktiengesellschaft für kombinatorische Chemie, München, Germany.
Conflict of Interest
The authors declare no conflict of interest.
17746
References
- Freiberg C, Fischer HP, Brunner NA (2005) Discovering the mechanism of action of novel antibacterial agents through transcriptional profiling of conditional mutants. Antimicrob Agents Chemother 49: 749-759.
- Bandow JE, Brötz H, Leichert LI, Labischinski H, Hecker M (2003) Proteomic approach to understanding antibiotic action. Antimicrob Agents Chemother 47: 948-955.
- Wecke T, Mascher T (2011) Antibiotic research in the age of omics: from expression profiles to interspecies communication. J AntimicrobChemother 66: 2689-2704.
- Lee CR, Lee JH, Park KS, Jeong BC, Lee SH (2015) Quantitative proteomic view associated with resistance to clinically important antibiotics in Gram-positive bacteria: a systematic review. Front Microbiol 6: 828.
- Fouhy F, Stanton C, Cotter PD, Hill C, Walsh F (2015) Proteomics as the final step in the functional metagenomics study of antimicrobial resistance. Front Microbiol 6: 172.
- Rashid MU, Dalhoff A, Weintraub A, Nord CE (2014) In vitro activity of MCB3681 against Clostridium difficile strains. Anaerobe 28: 216-219.
- Dalhoff A, Rashid MU, Kapsner T, Panagiotidis G, Weintraub A ,et al. (2015) Analysis of effects of MCB3681, the antibacterially active substance of prodrug MCB3837, on human resident microflora as proof of principle. ClinMicrobiol Infect 21: 761-764.
- Gertz S, Engelmann S, Schmid R, Ohlsen K, Hacker J,et al.( 1999) Regulation of sigmaB-dependent transcription of sigB and asp23 in two different Staphylococcus aureus strains. Mol Gen Genet 261: 558-566.
- Herbert S, Ziebandt AK, Ohlsen K, Schäfer T, Hecker M ,et al. (2010) Repair of global regulators in Staphylococcus aureus 8325 and comparative analysis with other clinical isolates. Infect Immun 78: 2877-2889.
- Jean BP, Franklin RC (2009) Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically: approved standard.Clinical and Laboratory Standards Institute (CLSI).pp.1-86
- Engelmann S, Hecker M (2008) Proteomic analysis to investigate regulatory networks in Staphylococcus aureus. Methods MolBiol 431: 25-45.
- Reiss S, Pane-Farre J, Fuchs S, Francois P,Liebeke M, et al. (2012) Global analysis of the Staphylococcus aureus response to mupirocin. Antimicrob Agents Chemother 56:787-804.
- Büttner K, Bernhardt J, Scharf C, Schmid R, Mäder U, et al. (2001) A comprehensive two-dimensional map of cytosolic proteins of Bacillus subtilis. Electrophoresis 22: 2908-2935.
- Voigt B, Schweder T, Becher D, Ehrenreich A, Gottschalk G, et al. (2004) A proteomic view of cell physiology of Bacillus licheniformis. Proteomics 4: 1465-1490.
- Wolf C, Hochgräfe F, Kusch H, Albrecht D, Hecker M, et al. (2008) Proteomic analysis of antioxidant strategies of Staphylococcus aureus: diverse responses to different oxidants. Proteomics 8: 3139-3153.
- Bernhardt J, Büttner K, Scharf C, Hecker M (1999) Dual channel imaging of two-dimensional electropherograms in Bacillus subtilis. Electrophoresis 20: 2225-2240.
- Cosgrove K, Coutts G, Jonsson IM, Tarkowski A, Kokai-Kun JF, et al. (2007) Catalase (KatA) and alkyl hydroperoxidereductase (AhpC) have compensatory roles in peroxide stress resistance and are required for survival, persistence, and nasal colonization in Staphylococcus aureus. J bacteriol 189: 1025-1035.
- Gray C, Moes L, Locher H, Dale G (2005) Characterization of MCB3681, a dual-action antibiotic. 45th ICAAC: Abstract No F-51.
- Bonn F, Pane-Farre J, Schlüter R, Schaffer M,Fuchs S, et al. (2016) Global analysis of the impact of linezolid onto virulence factor production in S. aureus USA300. Int J Med Microbiol 306: 131-140.
- Henze U, Sidow T, Wecke J, Labischinski H, Berger-Bächi B (1993) Influence of femB on methicillin resistance and peptidoglycan metabolism in Staphylococcus aureus. J Bacteriol 175: 1612-1620.
- CirzRT, Jones MB, Gingles NA, Minogue TD, Jarrahi B, et al. (2007) Complete and SOS-mediated response of Staphylococcus aureus to the antibiotic ciprofloxacin. J bacteriol 189: 531-539.






