Keywords
HIV seropositive; Multidrug resistance; Plasmid profile; ERIC-PCR; MAR
Introduction
Studies have shown that bacterial infections represent an important cause of morbidity and mortality in HIV infected patients [1-3]. The predominant causative organisms are the encapsulated bacteria, notably Streptococcus pneumoniae and Haemophilus influenza; but non-typhoidal Salmonella, Staphylococcus aureus and Pseudomonas aeruginosa have also been implicated [4-6].
S. aureus has been reported to colonize the anterior nares of HIV-infected patients with a higher frequency than the nares of HIV negative individuals, suggesting the possibility that high colonization burden may lead to a higher incidence of infection [6-8]. P.aeruginosa is notable for its multiple antibiotic resistance and is recognized as one of the pathogens that cause a variety of health care-associated infections [9]. It has been associated with infections of the urinary tract, respiratory system, soft tissue infection, gastrointestinal infection, dermatitis, cystic fibrosis and burns. The organism is also considered an opportunistic pathogen primarily because of its ability to undermine the immune mechanisms of immunocompromised HIV patients and those with impaired homeostasis mechanisms [10-13]. E. coli is a member of the normal flora of the digestive tract and its various serotypes are associated with different infections in humans [14]. One of these serotypes is the Shiga toxin producing and highly virulent E. coli O157: H7 which causes hemorrhagic colitis, hemolytic uremic syndrome and thrombocytopenic purpura [15,16]. In this study, we report the patterns of colonization and the characterization of the antibiotic resistance gene profiles of E.coli, S. aureus, P. aeruginosa and P.flourescens that were isolated from HIV seropositive and seronegative individuals in Akure, Southwestern Nigeria. It is expected that the results obtained in this study would elucidate bacterial colonization patterns among HIV seropositive patients and improve the antibiotic management of opportunistic infections in such individuals.
Materials and Methods
Study area and sample collection
The study was undertaken at the State Specialist Hospital, Akure (Ondo State, Southwestern Nigeria) with a population of 387,100 inhabitants. A total of 121 participants from the HIV clinic of the hospital were recruited for this study. They included 70 HIV seropositive patients and 51 HIV seronegative individuals. The HIV seropositive patients consisted of 52 females [mean age 36.8 years] and 18 males [mean age 45 years], while the HIV seronegative individuals comprised of 36 females [mean age 31.6 years] and 15 males [mean age 32.5 years]. Ethical clearance was obtained from both the State Specialist Hospital Management Board and the Ethical Review Board. Throat, rectal and skin swabs were collected using sterile cotton-tipped applicators (Evepon, Nigeria) dipped in sterile physiological saline. The skin swabs were collected from the elbow skin of the left hand (5 cm radius) from each patient for the sake of uniformity. Each swab was inoculated into fluid thioglycolate broth (Oxoid, England) for 24 hour growth at 37°C. Thereafter the cultures were processed and the bacterial isolates identified using Gram stain and biochemical procedures [17-19].
Antibiotics sensitivity tests
Selected isolates were tested for their susceptibility to commonly prescribed antibiotics by the Kirby Bauer disc diffusion method [20-22]. The list of antibiotics included beta-lactams, aminoglycosides, macrolides, hydroquinolones, trimethoprim and vancomycin. Multi-drug resistance among the isolates was defined as resistance to ≥ 1 agent in ≥ 3 antibiotic classes. S.aureus ATCC 25923 and Enterobacter aero-genes (American Type Culture Collection, Rockville, USA) were used as control organisms.
Molecular Characterization of the Bacteria Isolates
DNA extraction and plasmid profiling
DNA extraction for each bacterial culture was carried out by two cycles of boiling, homogenization, vortexing and centrifugation [23]. The purity and concentration of the DNAcontaining supernatant were determined using a Nanodrop spectrophotometer. The extraction of plasmid DNA from an overnight culture of each bacterial isolate was carried out by the TENS – Mini preparation method with Tris 25mM, EDTA 10mM,NaOH 0.1N and SDS 0.5% [24] in 5 ml of nutrient broth, followed by centrifugation, vortexing and pellet extraction in broth. About 300 μl of TENS solution was added, mixed by inversion 3-5 times until the solution became slimy after which 150 μl of 3.0M sodium acetate (pH 5.2) was added, vortexed for about 10 seconds and centrifuged at 13,000 rpm for 5 minutes. The supernatant was transferred into another eppendorf tube, and 900 μl of ice cold absolute ethanol was added, vortexed, and centrifuged for 10 minutes at 13,000 rpm. The supernatant was discarded, and the plasmid DNA pellet was rinsed twice with 1000 μl of 70% ethanol by centrifuging at 13,000 rpm for 5 minutes, and discarding the supernatant. The pellet was then air-dried, and 40 μl of TE buffer (Tris 10mM; 1mM Na2 EDTA) was added to re-suspend the pellet. Each bacterial isolate was run on 0.8% agarose gel electrophoresis at 80V for 90 minutes, using HIND III digest of Lambda DNA (Fermentas, USA) as the molecular weight marker.
Enterobacterial Repetitive Intergenic Consensus -Polymerase Chain Reaction Assay (ERIC-PCR)
ERIC-PCR was used to assess the clonal relatedness and diversity of the isolates. Two primers; forward primer ERIC 1 (5’-ATGTAAGCTCCTGGGGATTCAC-3’) and reverse primer ERIC 2 (5’AAGTAAGTGACTGGGGTGAGCG-3’) (Solis Biodyne) [25- 27] were used to amplify repetitive sequences present in the chromosomal DNA of the isolates. ERIC-PCR was carried out in 25 μl volume containing 2.5 unit of Taq DNA polymerase, 200 μM deoxynucleoside triphosphates (dNTPs), 2.5 mM MgCl2, 1X PCR Buffer, 100 ng of DNA and 20 pmol of each forward and reverse primer (Solis Biodyne) in sterile deionized water. Sample was amplified in an Eppendorf Nexus Thermal cycler [23] with an initial denaturation at 95°C for 3 minutes, then 35 cycles of denaturation at 95°C for 30 seconds, annealing at 52°C for 30 seconds, and extension at 72°C for 30 seconds. This was followed by a final extension step of 10 minutes at 72°C. The digests were then run on 1.5% agarose gel electrophoresis and visualized by ethidium bromide staining under UV trans-illuminator.
Approximately 1kb DNA ladder (Solis Biodyne) was used as DNA molecular weight standard.
Molecular identification of isolates
The isolates were subjected to PCR assay using specie specific primers to confirm phenotypic identification of the isolates. Pseudomonas aeruginosa specie-specific primer (forward primer 5-GGCGTGGGTGTGGAAGTC-3 and reverse primer 5- TGGTGGCGATCTTGAACTTCTT-3), Staphylococcus aureus specie-specific primer (Staph 756 forward primer 5-AACTCTGTTATTAGGGAAGAACA-3 and Staph 750 reverse primer 5-CCACCTTCCTCCGGTTTGTCACC-3), and Escherichia coli speciespecific primer (forward primer 5-AGAGCGCGAGATTATCAAGG-3 and reverse primer 5-TGCAGAGGCGAAGAAGTAAG-3) were used to amplify 16S rRNA segments of each respective bacterial specie. The cycling parameters for each specie was as follows: for Pseudomonas sp., initial denaturation was carried out at 94°C for 5 minutes, followed by 30 cycles of denaturation at 94°C for 1 minute, annealing at 55°C for 1 minute, and extension at 72°C for 1 minute, with a final extension step of 72°C for 7 minutes. For S. aureus, initial denaturation was also carried out at 95°C for 5 minutes, followed by 35 cycles of denaturation at 95°C for 30 seconds, annealing at 55°C for 1 minute, extension at 72°C for 90 seconds and a final extension of 4 minutes at 72°C. For E. coli, initial denaturation at 95°C for 3 minutes, 30 cycles of denaturation at 95°C for 30 seconds, 58°C for 30 seconds, and 72°C for 30 seconds, then final extension at 72°C for 5 minutes. The digests were then run on 1.5% agarose gel electrophoresis and visualized by ethidium bromide staining under UV transilluminator. 1kb DNA ladder (Solis Biodyne) was used as DNA molecular weight standard.
Detection of virulence and resistance genes by Polymerase Chain Reaction
Specific primers were also used to amplify sequences of the virulence and resistance genes. Details of primer sequences, predicted size of the amplified product and specific annealing temperature are given in Table 1.
| Organisms |
Target genes |
Primer sequence |
Amplicon size (bp) |
Annealing temp (°C) |
| Pseudomonas sp Virulence genes |
phzH
phzH |
F/ -5 GGGTTGGGTGGATTACAC-3
R/ -5 CTCACCTGGGTGTTGAAG-3 |
1,752 |
52 |
apr
apr |
F/ -5 TGTCCAGCAATTCTCTTGC -3
R/ -5 CGTTTTCCACGGTGACC -3 |
1,017 |
52 |
pvd
pvd |
F/ -5 GACTCAGGCAACTGCAAC -3
R/ -5 TTCAGGTGCTGGTACAGG -3 |
1,281 |
59 |
lasB
lasB |
F/ -5 ACAGGTAGAACGCACGGTTG-3
R/ -5 GATCGACGTGTCCAAACTCC-3 |
1,220 |
55 |
| Pseudomonas sp Resistance genes |
blaPSE |
F/ -5 ACCGTATTGAGCCTGATTTA -3
R/ -5 ATTGAAGCCTGTGTTTGAGC-3 |
321 |
60 |
| blaAMPC |
F/ -5 GGTATGGCTGTGGGTGTTA -3
R/ -5 TCCGAAACGGTTAGTTGAG -3 |
882 |
53 |
| blaIMP |
F/ -5 CTACCGCAGCAGAGTCTTGG -3
R/ -5 AACCAGTTCTGCCTTACCAT -3 |
Variable |
55 |
aadA
aadA |
F/ -5 CTTGATGAAACAAGGCGG -3
R/ -5 TACCAAATGCGGGACAAC -3 |
Variable |
55 |
| Staphylococcus aureus |
mecA1 |
F/ -5 AATATCGATGGTAAAGGTTGGC -3
R/ -5 AGTTCTGCAGTACCGGATTTGC-3 |
528 |
55 |
| Luk-pv |
F/ -5 ATCATTAGGTAAAATGTCTGGACATGATCCA -3
R/ -5 GCATCAAGTGTATTGGATAGCAAAAGC-3 |
|
55 |
Legend:F/ Forward primer: R/ Reverse primer
Table 1: The PCR primers sequences used for virulence and resistance factors
Data analysis
Multiple antibiotic resistance (MAR) index for each test isolate was calculated as proposed by Krumperman et al. [28]. The MAR index was determined by dividing the number of antibiotics to which test isolate displayed resistance by the total number of antibiotics against which the test organism has been evaluated for sensitivity. Data generated for age, weight, height and gender were also evaluated using independent t-test with P ≤ 0.05 as indicator of statistical significance. Windows SPSS version 16.0 was used to perform the analyses.
Results
A total of 96 bacterial isolates of E. coli, S. aureus, P. aeruginosa and P.fluorescens isolated from HIV seropositive individuals and seronegative controls were used in this study. The distribution of the bacterial isolates is shown on Table 2 and is also illustrated in Figure 1. These included 46 isolates from 70 HIV seropositive individuals and 50 isolates from 51 seronegative controls. None of the Pseudomonas specie analyzed in this study was from HIV seronegative controls. The results in Tables 3 and 4 show that 39 (84.8%) of bacterial isolates from HIV seropositive subjects were resistant to more than three classes of antibiotics as compared to 78% of isolates from seronegative individuals. Twenty (90.9%) of 22 Pseudomonas isolates were resistant to 3 or more classes of antibiotics and all were resistant to cefuroxime. All the P. aeruginosa isolates from HIV seropositive patients were resistant to ampicillin, oxacillin, cefoxitin, cefuroxime, cephalotin, vancomycin and erythromycin, while 91.7% of them were resistant to augmentin (amoxicillin with clavulanic acid), ceftriazone, kanamycin and trimethoprim. This high level of multidrug resistance, in addition to those of penicillin derivatives, augmentin and cephalosporins, suggests that the isolates were most likely extended spectrum beta lactamase producers [29]. Both Pseudomonas species are an important cause of various infections and have been associated with high rates of morbidity and mortality among patients with HIV infection [30]. The S. aureus isolates also exhibited some degree of multipleresistance. About 66.7% of the isolates were also resistant to 3 or more different classes of antibiotics. All the E. coli isolates were multi-resistant as they all had resistances to more than 3 classes of antibiotics.
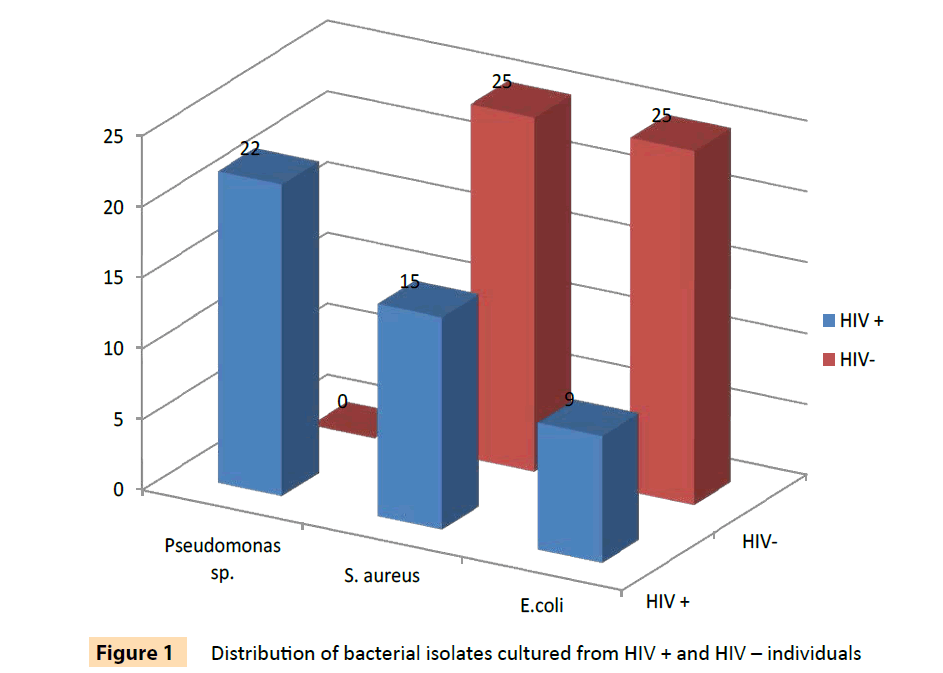
Figure 1: Distribution of bacterial isolates cultured from HIV + and HIV – individuals
| |
Pseudomonas sp. |
Staphylococcus aureus |
Escherichia coli |
Total |
| |
Skin |
Throat |
Rectal swab |
Skin |
Throat |
Rectal swab |
Skin |
Throat |
Rectal swab |
|
| HIV + |
9 |
8 |
5 |
2 |
9 |
4 |
4 |
1 |
4 |
46 |
| HIV- |
- |
- |
- |
4 |
15 |
6 |
5 |
2 |
18 |
50 |
| TOTAL |
9 |
8 |
5 |
6 |
24 |
10 |
9 |
3 |
22 |
96 |
HIV: human immunodeficiency virus
Table 2: The distribution of bacterial isolates obtained from HIV + and HIV- individuals in Akure, Southwestern Nigeria
| ANTIBIOTICS |
Pseudomonas sp. |
Staphylococcus aureus |
Escherichia coli |
| HIV + |
HIV- |
HIV + |
HIV- |
HIV + |
HIV- |
| |
n= 22 (%) |
n= 0 (%) |
n= 15 (%) |
n= 25 (%) |
n= 9 (%) |
n= 25 (%) |
| AMP |
20 (90.9) |
ND |
14 (93.3) |
21 (84.0) |
9 (100.0) |
13 (52.0) |
| OXA |
21 (95.5) |
ND |
9 (60.0) |
6 (24.0) |
9 (100.0) |
25 (100.0) |
| AMC |
18 (81.8) |
ND |
9 (60.0) |
4 (16.0) |
5 (55.6) |
1 (4.0) |
| FOX |
21 (95.5) |
ND |
15 (100.0) |
10 (40.0) |
9 (100.0) |
3 (12.0) |
| CAZ |
20 (90.9) |
ND |
13 (86.7) |
5 (20.0) |
9 (100.0) |
1 (4.0) |
| CRO |
20 (90.9) |
ND |
11 (73.3) |
8 (32.0) |
8 (88.9) |
3 (12.0) |
| CXM |
22 (100.0) |
ND |
14 (93.3) |
3 (12.0) |
9 (100.0) |
1 (4.0) |
| KF |
21 (95.5) |
ND |
11 (73.3) |
4 (16.0) |
9 (100.0) |
18 (72.0) |
| IPM |
1 (4.5) |
ND |
2 (13.3) |
0 (0.0) |
0 (0.0) |
0 (0.0) |
| VAN |
19 (86.4) |
ND |
7 (46.7) |
3 (12.0) |
7 (77.8) |
25 (100.0) |
| AMK |
8 (36.4) |
ND |
6 (40.0) |
1 (4.0) |
5 (55.6) |
0 (0.0) |
| GEN |
1 (4.5) |
ND |
2 (13.3) |
2 (8.0) |
0 (0.0) |
0 (0.0) |
| KAN |
17 (77.3) |
ND |
4 (26.7) |
6 (24.0) |
3 (33.3) |
1 (4.0) |
| STR |
12 (54.5) |
ND |
4 (26.7) |
1 (4.0) |
6 (66.7) |
13 (52.0) |
| ERY |
18 (81.8) |
ND |
4 (26.7) |
5 (20.0) |
6 (66.7) |
21 (84.0) |
| CIP |
5 (22.7) |
ND |
5 (33.3) |
3 (12.0) |
4 (44.4) |
1 (4.0) |
| OFX |
7 (31.8) |
ND |
4 (26.7) |
1 (4.0) |
1 (11.1) |
1 (4.0) |
| TRI |
20 (90.9) |
ND |
10 (66.7) |
20 (80.0) |
9 (100.0) |
17(68.0) |
Legend:AMP (Ampicillin 10μg), OXA (oxacillin 1μg), AMC (Amoxicillin/clavulanic acid 20μg), FOX (Cefoxitin 30μg), CAZ (Ceftazidime 30μg), CRO ((Ceftriaxone 30μg), CXM (Cefuroxime 30μg), KF (Cephalotin 30μg), IPM (Imipenem 10μg), VAN (Vancomycin 30μg), AMK (Amikacin 30 μg), GEN (Gentamicin10μg), KAN (Kanamycin 30μg), STR (Streptomycin 10μg), ERY (Erythromycin 15μg), CIP (Ciprofloxacin 5μg), OFX (Ofloxacin5 μg), TRI (Trimethoprim5μg).
Table 3: The Antibiotic Resistance profile of bacterial isolates obtained from HIV + and HIV- individuals in Akure, Southwestern Nigeria.
| |
NO OF ANTIBIOTIC CLASS TO WHICH ISOLATES ARE RESISTANT |
| HIV POSITIVE |
NO TESTED |
0 |
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
| Staphylococcus aureus |
15 |
0 |
0 |
5 |
0 |
2 |
2 |
4 |
1 |
1 |
| Escherichia coli |
9 |
0 |
0 |
0 |
0 |
1 |
3 |
3 |
2 |
0 |
| Pseudomonas sp. |
22 |
0 |
0 |
2 |
0 |
2 |
1 |
11 |
5 |
1 |
| Total |
46 |
|
|
|
|
|
|
|
|
|
| HIV NEGATIVE |
|
|
|
|
|
|
|
|
|
|
| Staphylococcus aureus |
25 |
1 |
5 |
6 |
1 |
4 |
7 |
1 |
0 |
0 |
| Escherichia coli |
25 |
0 |
0 |
0 |
1 |
14 |
7 |
3 |
0 |
0 |
| Total |
50 |
|
|
|
|
|
|
|
|
|
Table 4: The Multiple Antibiotic Resistance pattern of bacterial isolates
Randomly selected 10 isolates of P. aeruginosa, eight P. fluorescens isolates, nine E. coli isolates and three S. aureus isolates from HIV seropositive patients as well as 14 E. coli and 10 S. aureus isolates from seronegative controls were subjected to plasmid profiling and genotyping using ERIC-PCR. The results in Table 6 revealed ten band patterns for the Pseudomonas isolates, two band patterns for E. coli isolates and four band patterns for seven of the S. aureus isolates. The variations in band patterns suggest the existence of strains within the four bacterial species. Figures 2-6b also show that eight of the 18 Pseudomonas isolates carried the blaPSE gene, while four had the aadA gene, although none of them carried a single plasmid. One of the isolates also carried both blaPSE and aadA genes. In contrast, only two of the eight S. aureus isolates carried the mecA gene for methicillin resistance. Each of the two isolates came from a seropositive HIV patient and a seronegative HIV control. The results also show that the MAR indices in this study were >0.2 in 95.5% of Pseudomonas species, 86.7% of S. aureus and 100% for E. coli isolated from HIV seropositive patients (Table 5). The MAR indices were also > 0.2 in 40% and 64% for S. aureus and E. coli from HIV seronegative individuals respectively. These suggest that the environment from which the isolates were obtained is an antibiotic pressurized one.
| MAR index |
Pseudomonas sp. |
S. aureus |
E. coli |
| |
HIV + |
HIV- |
HIV + |
HIV- |
HIV + |
HIV- |
| |
n= 22 |
n= 0 |
n= 15 |
n= 25 |
n= 9 |
n= 25 |
| 0 |
0 (0.0) |
ND |
0 (0.0) |
1 (4.0) |
0 (0.0) |
0 (0.0) |
| 0.1 |
0(0.0) |
ND |
0(0.0) |
11 (44.0) |
0(0.0) |
0(0.0) |
| 0.2 |
1 (4.5) |
ND |
2 (13.3) |
3 (12.0) |
0(0.0) |
9 (36.0) |
| 0.3 |
1 (4.5) |
ND |
4 (26.7) |
2 (8.0) |
0(0.0) |
6 (24.0) |
| 0.4 |
1 (4.5) |
ND |
0(0.0) |
5 (20.0) |
0(0.0) |
8 (32.0) |
| 0.5 |
1 (4.5) |
ND |
1 (6.7) |
3 (12.0) |
1 (11.1) |
1 (4.0) |
| 0.6 |
1 (4.5) |
ND |
3 (20.0) |
0(0.0) |
2 (22.2) |
1 (4.0) |
| 0.7 |
11 (50.0) |
ND |
2 (13.3) |
0(0.0) |
4 (44.4) |
0(0.0) |
| 0.8 |
2 (9.1) |
ND |
0(0.0) |
0(0.0) |
2 (22.2) |
0(0.0) |
| 0.9 |
3 (13.6) |
ND |
2 (13.3) |
0(0.0) |
0(0.0) |
0(0.0) |
| 1.0 |
1 (4.5) |
ND |
1 (6.7) |
0(0.0) |
0(0.0) |
0(0.0) |
Legend: MAR-Multi-antibiotic resistance index
Table 5: The Multiple Antibiotic Resistance Index pattern of bacterial isolates
| Bacteria specie |
Variants |
Base pairs of the bands |
No of isolates |
| Pseudomonas sp. |
1 |
280, 300, 350, 400, 500 |
1 |
| 2 |
280, 300, 480, 500 |
2 |
| 3 |
280, 300, 350, 480 |
1 |
| 4 |
280, 300, 350, 400 |
3 |
| 5 |
300, 350, 400, 500 |
1 |
| 6 |
280, 320, 480 |
1 |
| 7 |
150, 300, 400 |
1 |
| 8 |
150, 300 |
1 |
| 9 |
350 |
2 |
| 10 |
300 |
2 |
| Escherichia coli |
1 |
220, 280 |
6 |
| 2 |
150, 280 |
1 |
| Staphylococcus aureus |
1 |
100, 150, 250, 290 |
7 |
| 2 |
100, 250, 290 |
1 |
| 3 |
150, 250, 290 |
1 |
| 4 |
100, 250 |
4 |
Table 6: Probable variants of bacterial isolates subjected to ERIC-PCR using ERIC 1 and ERIC 2 primers
Figure 2: The gel electrophoresis PCR products using specie specific primers for the identification of Escherichia coli
Legend: Lane M: Molecular weight marker; Lanes 1-9E. coli isolates cultured from HIV seropositive patients. Lanes 11 – 24 - E. coli isolates cultured from HIV seronegative individuals
Figure 3: The gel electrophoresis of PCR products using specie specific primers for the identification of Staphylococcus aureus isolated from HIV positive individuals Akure, Ondo State, Nigeria
Legend: Lane M: Molecular weight marker; Lanes 1-3S. aureusisolates cultured from HIV seropositive patients. Lanes 4 - 13S. aureus isolates cultured from HIV seronegative individuals
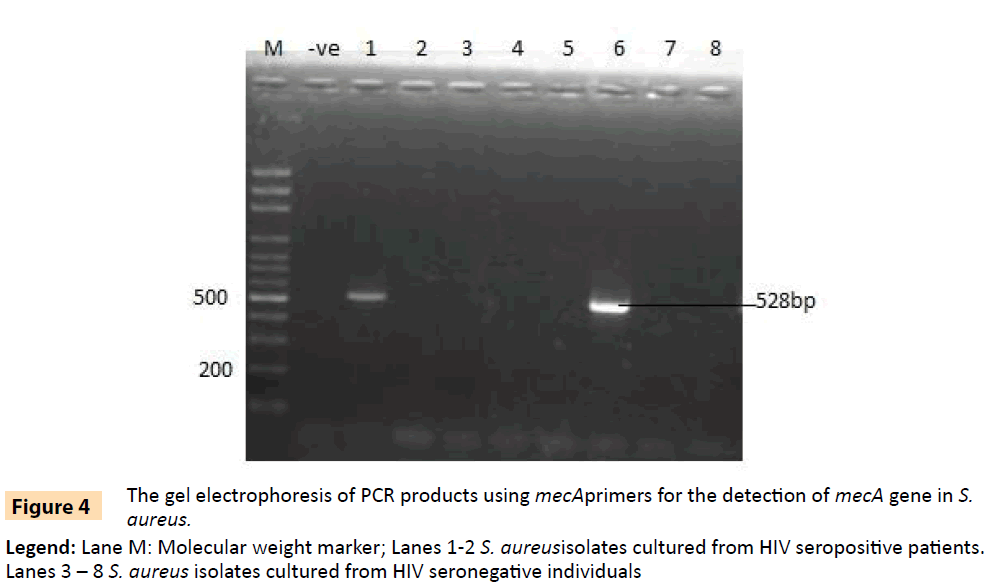
Figure 4: The gel electrophoresis of PCR products using mecAprimers for the detection of mecA gene in S. aureus.
Legend: Lane M: Molecular weight marker; Lanes 1-2 S. aureusisolates cultured from HIV seropositive patients. Lanes 3 – 8 S. aureus isolates cultured from HIV seronegative individuals
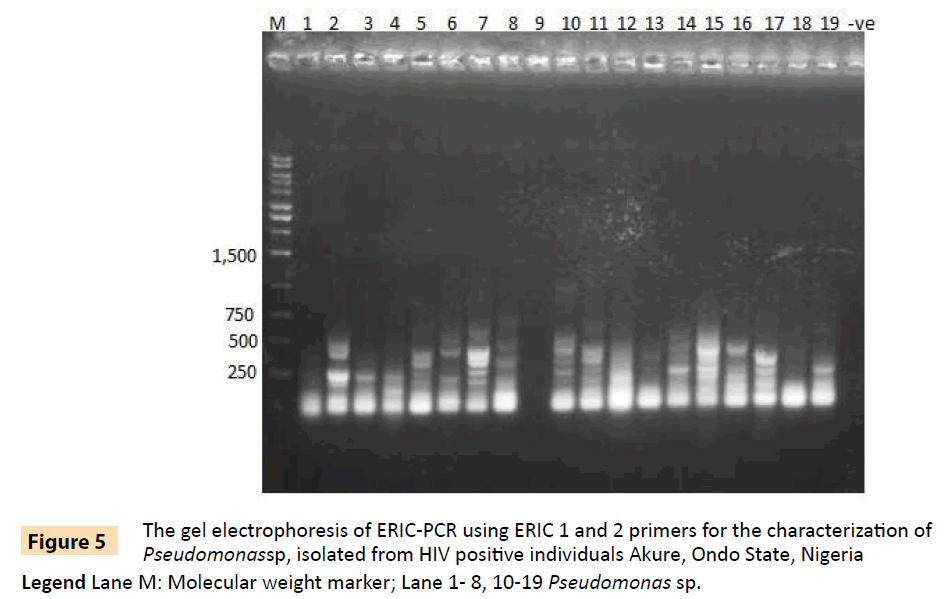
Figure 5: The gel electrophoresis of ERIC-PCR using ERIC 1 and 2 primers for the characterization of Pseudomonassp, isolated from HIV positive individuals Akure, Ondo State, Nigeria.
Legend Lane M: Molecular weight marker; Lane 1- 8, 10-19 Pseudomonas sp.
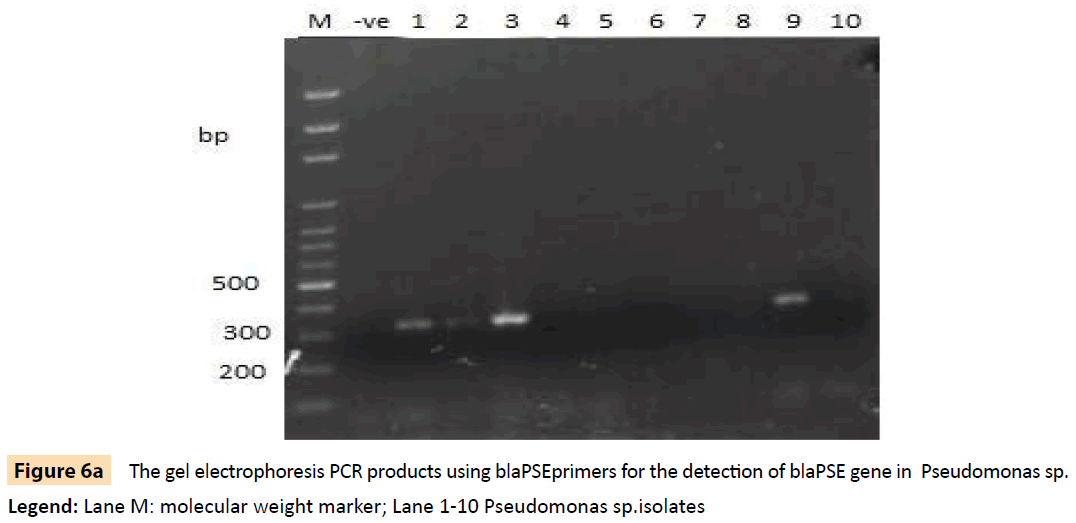
Figure 6a: The gel electrophoresis PCR products using blaPSEprimers for the detection of blaPSE gene in Pseudomonas sp.
Legend: Lane M: molecular weight marker; Lane 1-10 Pseudomonas sp.isolates
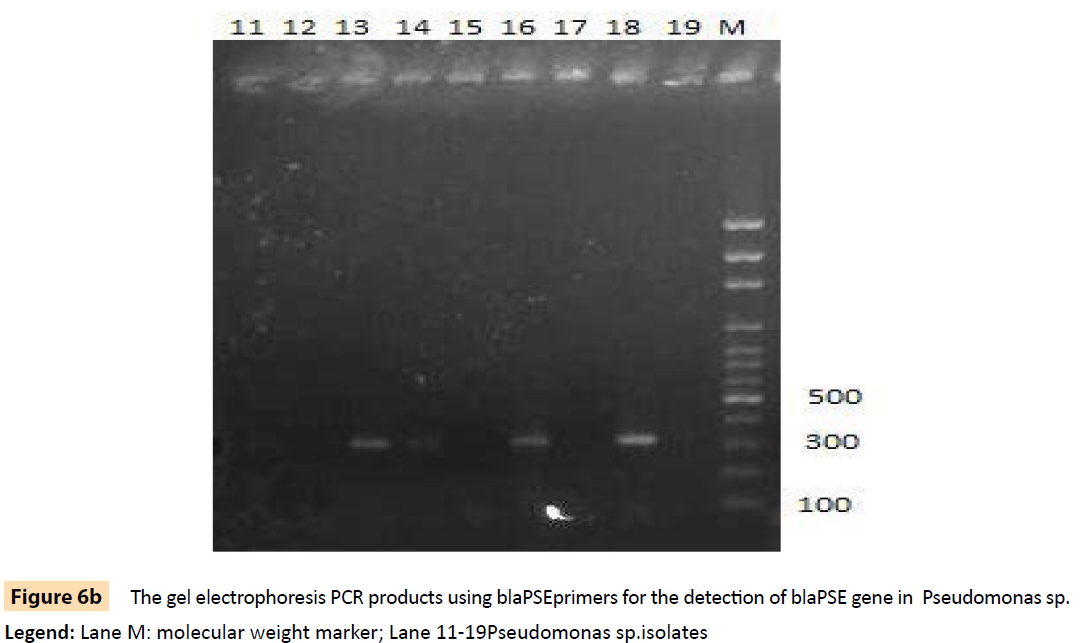
Figure 6b: The gel electrophoresis PCR products using blaPSEprimers for the detection of blaPSE gene in Pseudomonas sp.
Legend: Lane M: molecular weight marker; Lane 11-19Pseudomonas sp.isolates
Discussion
We have shown in this study that HIV seropositive patients have a higher degree of colonization with multiple antibiotic resistant E.coli, S. aureus, P. aeruginosa and P. flourescens. While we did not isolate a single plasmid from any of the 96 bacterial isolates, we showed that several of the isolates carried resistance and virulence genes [31-36]. Twenty eight (60.9%) of the isolates were found to be resistant to at least six different classes of antibiotics. Results of the ERIC-PCR showed ten (10) gene band patterns for the two Pseudomonas species, four band patterns for S. aureus and two patterns for E. coli, suggesting a diversity of strains among the isolates. Four of the 18 Pseudomonas isolates carried aadA gene while eight had the blaPSE gene (Table 6). Only two isolates out of the eight of S. aureus isolates that were tested carried the mecA gene. The MAR index of the tested isolates revealed that 93.5% of the isolates from HIV seropositive individuals were above 0.2, suggesting an antibiotic pressurized community. Colonization with S. aureus has been reported to be a risk factor for subsequent clinical infections in HIV positive patients [37,38]; and the site of colonization may also play a key risk factor [39]. The anterior nares as well as the skin, throat, ear and feet are considered a major reservoir of S. aureus in this environment [40]. The presence of such multi-resistant strains on the body surface of immune-compromised individuals portends serious implications in view of the fact that these organisms have also been known to cause recurrent infections [3]. Globally, Staphylococcus aureus infections have been reported to be important causes of morbidity and mortality [41]. HIV positive individuals are at increased risk of opportunistic and common bacterial infections, and S. aureus ranks as one of the most common causes of bacterial infections [42-44]. Furthermore, HIV infection is associated with a higher risk of recurrent infection [45]. Interestingly, despite the high degree of resistance among isolates from HIV seropositive patients, some of the isolates were still sensitive to imipenem as well as gentamycin. This corroborates previous studies in which investigators reported antibiotic resistance patterns of Pseudomonas aeruginosa to imipenem. Resistance patterns to imipenem has been found to be lower than to other antibiotics; 16.3% in Brazil [46], 2.9% in Iran [47], 21% in Greece [48] and 13% in Spain [49]. All these studies reported low resistance values to imipenem. The low level of resistance to gentamycin has also been recorded in other studies [50-53], and it has been suggested in all cases that the efficacy of gentamycin may be related to the mode of its administration which has limited its abuse and misuse.
The study also revealed the presence of resistance genes (mecA, blaPSE and aadA) in some of the bacterial isolates analyzed from the HIV seropositive as well as HIV seronegative individuals. About 90.9% of the two Pseudomonas species were multi-resistant to more than 3 classes of antibiotics among seropositive patients. The expression of the efflux pump has been implicated in resistance of Pseudomonas sp. to beta lactam antibiotics. These proteins transport the antibiotics from within the cell to the external environment. This multi- drug system has been shown to provide resistance to a very wide range of compounds in Gram negative bacilli [31,32], coupled with a low permeability of the outer membrane [33], and a remarkable ability to acquire further resistance mechanisms to multiple groups of antimicrobial agents, including β-lactams, aminoglycosides and fluoroquinolones [34]. It is notable that many of the resistant mechanisms are often present simultaneously, thereby conferring multi-resistant properties to the organism [35]. Studies have shown that P. aeruginosa is the second most common cause of nosocomial pneumonia (17%), the third most common cause of urinary tract infection (7%), the fourth most common cause of surgical site infection (8%), the seventh most frequently isolated pathogen from the bloodstream (2%) and the fifth most common isolate (9%) overall from all sites [36].
MAR values calculated from this study show that the indices for the HIV positive population were higher than those of their HIV negative controls. MAR indexing is an effective as well as a useful tool for evaluating the spread of bacterial resistance in a given population [52]. The high MAR indices of organisms isolated from our HIV seropositive cohort poses a challenge to the clinical management of these immunocompromised individuals.
Antimicrobial resistance is an increasing problem worldwide. In Nigeria where health institutions have little-to-none antibiotic policy, widespread antibiotic resistance is common and seems higher in immune-compromised subjects than in immunecompetent individuals as revealed by our data. Therefore, there is still an urgent need for continuous and constant monitoring of resistance patterns in bacteria in view of the increasing emergence of multidrug resistance among different bacterial species. Our findings have therefore provided baseline data to institute effective therapeutic strategies in combating the challenges of antibiotic resistance among healthcare providers in Nigeria. Hence, surveillance data, continuous antibiotic susceptibility testing in hospitals and treatment regimens based on history of previous antibiotics use as well as education of the population may lower the incidence of multidrug resistant organisms.
3808
References
- Berger,BJ.,Hussain, F., Roistacher, K. Bacterial infections in HIV-infected patients. Infect Dis Clin N Am1994; 8: 449-465.
- Schuster,MG., Norris, AH. Community-acquired Pseudomonas aeruginosapneumonia in patients with HIV infection. AIDS 1994; 8: 1437-1441.
- Domingo, P., Ferre, A, Baraldes, MA.,Ris, J., Sanchez F.Pseudomonas aeruginosabronchopulmonary infection in patients with AIDS, with emphasis on relapsing infection. EurRespirJ1998; 12: 107-112.
- Witt,DJ., Craven, DE., McCabe, WR. Bacterial infections in adult patients with the acquired immune deficiency syndrome (AIDS) and AIDS related complex. Am J Med1987; 82: 900-906.
- Onorato, M., Borucki, MJ., Baillargeon, G., Paar, DP., Freeman, DH., et al. Risk factors for colonization or infection due to methicillin-resistant Staphylococcus aureusin HIV-positive patients: a retrospective case-control study. Infect Control HospEpidemiol1999; 20: 26-30.
- Alicia,HI.,Kempker, R., Moann, A., Rimland, D., (2010). Methicillin-resistant Staphylococcus aureusin HIV-infected patients. Infection and Drug Resistance2010; 3: 73-86.
- Raviglione, MC., Mariuz, P., Pablos-Mendez, A., Battan, R., Ottuso, P., et al. High Staphylococcus aureusnasal carriage rate in patients with acquired immunodeficiency syndrome or AIDS-related complex. Am J Infect Control 1990; 18: 64-69.
- Weinke, T., Schiller, R., Fehrenbach,FJ.,Pohle, HD. (1992). Association between Staphylococcus aureusnasopharyngeal colonization and septicemia in patients infected with the human immunodeficiency virus. Eur J ClinMicrobiol Infect Dis 1992; 11: 985-989.
- Boucher,HW., Talbot, GH., Bradley, JS., Edwards, JE., Gilbert, D., et al.Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis 2009; 48:1-12.
- Walker, TS.Microbiology. O.W.B. Saunders, Philadelphia, PA 1998; 173-181.
- Kiska,DL., Gilligan, PH. Pseudomonas. In: Manual of Clinical Microbiology, Murray, P.R., E.J. Baron, M.A. Pfaller, F.C. Tenover and R.H. Yolken (Eds.). (7thedtn), ASM Press, Washington, DC.,1999; 517-525.
- Rowe, SM., Miller, S., Sorscher,EJ.,Cystic fibrosis. N Engl J Med 2005; 352: 1992-2001.
- Driscoll,JA., Brody, SL., Kollef, MH.The epidemiology, pathogenesis and treatment of Pseudomonas aeruginosa infections. Drugs 2007; 67: 351-368.
- Belanger, SD.,Boissinot, M., Menard, C., Picard,FJ., Bergeron, MJ. Rapid detection of Shiga toxin-producing bacteria in feces by multiplex PCR with molecular beacons on the Smart Cycler. JClinMicrobiol 2002;40: 1436-1440.
- Eliott, E., Robins-Browne, R., O'Loughlin, E., Bennett-Wood, V., Bourke, J.Nationwide study of haemolyticuraemic syndrome: clinical, microbiological, and epidemiological features. Arch Dis Child2001; 85: 125-131.
- Paton, AW., Paton, JC. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA,rfb0111, and rfb0157. J ClinMicrobiol 1998; 36:598-602.
- Cowan, S., Steel, J.Manual for the identification of Medical Bacteria. Cambridge University Press, 1975; 144.
- Koneman,EW., Allen, SD., Janda, WM., Schreckenbergr, PC., Winn, WC. Antimicrobial susceptibility testing, chapter 15. In: color Atlas and Textbook of Diagnostic Microbiology(5thedtn)Lippicott, Philadelphia 1997;785.
- Baron, E.,Finegold, S.Bailey and Scott’s diagnostic microbiology. St. Louis: Mosby, 1990; 861.
- Ericsson,HM.,Sherris, JC. The agar dilution method. ActaPatholMicrobiolScand B 1971; 217: 11.
- Bauer, A., Kirby,WW.,Sherris, JC., Turck, M. Antibiotic susceptibility testing by a standardized single disc method. Am J Clin Path 1966; 45: 493-496.
- CLSI (Clinical and Laboratory Standards Institute) Performance standards for antimicrobial disk susceptibility tests, (9thedtn). Approved Standard M2-A9. 2007; Wayne, PA: document M100-S17.
- Adeleke, MA.,Olaitan, JO., Abiona, O., Canice, J., Olajide, S., et al. Molecular Characterization And Antibiotic Susceptibility Patterns Of Bacteria Isolated From Wara(West African Cheese) Sold In Osun State, Nigeria. Innovative Romanian Food Biotechnology2014; 15: 23.
- Oleghe, P., Odimegwu, D., Udofia, E., Esimore, C. Multidrug resistant bacteria isolates recovered fromherbal medicinal preparations in Southeastern Setting, Nigeria. In Journal of Rural and Tropical Public Health 2011; 10: 70-75.
- Szcuka, E.,Kaznowski, A. Typing of clinical and environmental Aeromonas sp. strains by random amplified polymorphic DNA PCR, repetitive extragenic palindromic PCR, and enterobacterial repetitive intergenic consensus sequence PCR. J. Clin. Microbiol 2004; 42:220-228.
- Versalovic, J., Koeuth, T., Lupski, JR. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res1991; 19: 6823-6831.
- Hulton,CSJ., Higgins, CF., Sharp, PM. ERIC Sequences: a novel family of repetitive elements in the genomes of Escherichia coli, Salmonella typhimuriumand other Enterobacteria. MolMicrobiol 1991; 5: 825-834.
- Krumpermam, PH. Multiple antibiotics resistanceindexing of E. coli to identify high risks sources offaecal contamination of foods. App EnvMicrobiol1983; 46: 165-170.
- Bert, F., Branger, C., Lambert-Zechovsky, N. Identification of PSE and OXA ß-lactamase genes in Pseudomonas aeruginosa using PCR–restriction fragment length polymorphism. JAntimicrobChemother 2002; 50: 11-18.
- Manfredi, R., Nanetti, A., Ferri, M., Chiodo, F. Pseudomonas spp. complications in patients with HIV disease: an eight-year clinical and microbiological survey. Eur J Epidemiol 2000; 16:111-8.
- Mesaros, N., Nordmann, P., Plesiat, P., Roussel-Delvallez, M., Van Eldere, J., et al. Pseudomonas aeruginosa: resistance and therapeutic options at the turn of the new millenium.ClinMicrobiol Infect2007; 13;560-578.
- Poole, K. Efflux mediated multiple resistance in Gram negative bacteria. Clinical Microbiology and Infection 2004;10: 12-16.
- Livermore, DM.Penicillin-binding proteins, porins and outer-membrane permeability of carbenicillin-resistant and -susceptible strains of Pseudomonas aeruginosa. J Med Microbiol1984; 18: 261-270.
- Bubonja-Sonje, M., Matovina, M., Skrobonja, I., Bedenic, B., Abram, M. Mechanisms of Carbapenem Resistance in Multidrug-Resistant Clinical Isolates of Pseudomonas aeruginosa from a Croatian Hospital. Microbial drug resistance2015; Larchmont, NY.
- McGowan, JE Jr.(2006).Resistance in nonfermenting gram-negative bacteria: multidrug resistance to the maximum.Am J Infect Control2006; 34:S29-S37.
- National Nosocomial Infections Surveillance (NNIS) system report: data summary from January 1992 through June 2003, issued August 2003.
- Philip,PJ., John, T., Brooks, Sigrid, K., McAllister, et al.Methicillin-Resistant Staphylococcus aureus Colonization of the Groin and Risk for Clinical Infection among HIV-infected Adults. Emerging Infectious Diseases2013; 19: 4.
- Von Eiff, C., Becker, K., Machka, K., Stammer, H., Peters, G. Nasal carriage as a source of Staphylococcus aureusbacteremia. N Engl J Med2001; 344:11-6.
- Szumowski, JD., Wener, KM., Gold, HS., Wong, M., Venkataraman, L., et al. Methicillin-resistant Staphylococcus aureuscolonization, behavioral risk factors, and skin and soft-tissue infection at an ambulatory clinic serving a large population of HIV infected men who have sex with men. Clin Infect Dis 2009; 49: 118–21.
- Kwashie, ANA.,Muibat, AF., Modupe, AI., Adejumoke, BM., Kehinde, AO. A survey of bacterial isolates cultured from apparently healthy individuals in South-Western Nigeria. International Journal of Tropical Medicine2012; 7: 130-137.
- Bearman, GM., Wenzel, RP. Bacteremias: a leading cause of death. Arch Med Res 2005; 36: 646-659.
- Tumbarello, M., Tacconelli, E., Caponera, S., Cauda, R., Ortona L. The impact of bacteraemia on HIV infection. Nine years experience in a large Italian university hospital. J Infect 1995; 31: 123-131.
- Senthilkumar, A., Kumar, S., Sheagren, JN. Increased incidence of Staphylococcus aureus bacteremia in hospitalized patients with acquired immunodeficiency syndrome. Clin Infect Dis 2001; 33: 1412-1416.
- Laupland,KB., Church, DL., Mucenski, M., Sutherland, LR., Davies, HD. Population-based study of the epidemiology of and the risk factors for invasive Staphylococcus aureus infections. J Infect Dis 2007; 187: 1452-1459.
- Kreisel, K., Boyd, K., Langenberg, P., Roghmann, MC. Risk factors for recurrence in patients with Staphylococcus aureus infections complicated by bacteremia. DiagnMicrobiol Infect Dis2006; 55: 179-184.
- deFreitas, ALP., Barth AL. Antibiotic resistance and molecular typing of Pseudomonas aeruginosa: focus on imipenem. The Braz J Infect Dis 2002; 6: 1-7.
- Nikbin, VS., Abdi-Ali, A., Feizabadi,MM.,Gharavi, S. Pulsed-field gel electrophoresis and plasmid profile of Pseudomonas aeruginosaat two hospitals in Tehran, Iran. Indian J Med Res 2007; 126: 146-51.
- Tassios, PT., Gennimata, V., Maniatis, AN., Tassios, PT., Gennimata, V., et al. Emergence of multidrug resistance in ubiquitous and dominant Pseudomonas aeruginosaserogroup 0:11. J ClinMicrobiol 1998: 36: 897-901.
- Bouza, E., Garcia-Garrote, F., Cercenado, E., Marin, M., Diaz for the Spanish Pseudomonas aeruginosaStudy Group. Pseudomonas aeruginosa: a survey of resistance in 136 hospitals in Spain. Antimicrob Agents Chemother 1999; 43: 981-2.
- Ako-Nai,AK.,Adeyemi, FM., Aboderin, OA., Kassim, OO. Antibiotic resistance profile of staphylococci from clinical sources recovered from infants. African Journal of Biotechnology2005; 4: 816-822.
- Ako–Nai,AK.,Oluga, FA., Onipede, AO., Adejuyigbe, EA., Amusa, YB.The characterization of Bacterial Isolates from Acute OtitisMedia in Ile-Ife, Southwestern Nigeria. J Trop Pediatrics 2002: 48.
- Ehinmidu J. Antibiotics susceptibility patterns of urine bacteria isolates in Zaria, Nigeria. In Tropical Journal of Pharmaceutical Research 2003; 2: 223-228.
- Ako-Nai, AK.,Ebhodaghe, BI.,Osho, P., Adeyemi AFM.,Kassim, OO (2014). Preponderance of bacterial isolates in urine of HIV positive malaria infected pregnant women with urinary tract infection. Journal of Infection in Developing Countries8:1591-1600.












