Shilpa Rana1, Yusra Ahmad1, Narotam Sharma2*, Vijay Kumar2 and Satish Chandra Nautiyal2
1Faculty of Pharmacy, Uttarakhand Technical University, Suddhowala, Dehradun, Uttarakhand, India
2Central Molecular Research Laboratory, Department of Biochemistry, Shri Guru Ram Rai Institute of Medical and Health Sciences, Shri Guru Ram Rai University, Dehradun, Uttarakhand, India
- Corresponding Author:
- Narotam Sharma
Central Molecular Research Laboratory
Department of Biochemistry
Shri Guru Ram Rai Institute of Medical and Health Sciences
Shri Guru Ram Rai University, Dehradun, Uttarakhand, India
Tel: +919557650069
E-mail: sharmanarotam5@gmail.com
Received Date: April 16, 2019; Accepted Date: April 25, 2019; Published Date: May 03, 2019
Citation: Rana S, Ahmad Y, Sharma N, Kumar V, Nautiyal SC (2019) Molecular Characterization of Clinical Isolates to Study rpoB Gene in Mycobacterium tuberculosis Complex. Int J Drug Dev & Res 11: 30-33. doi: 10.36648/0975-9344.11.2.133
Copyright:© 2019 Rana S. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Keywords
PCR; MTB; mpb64 gene; rpoB gene; Rifampicin
Introduction
Antimicrobial resistance has become one of the major health problems, especially in developing countries. The easy availability and higher consumption of medicines lead to the emergence of drug resistance [1]. India has shown high burden of infectious diseases in the world and the recent report has shown inappropriate and irrational use of antimicrobial agents against these diseases. The report has also shown that a large population in India suffers from inadequacy of finance, due to which favourable condition for drug resistance occurs [2]. In India, a recent study was conducted and highlighted the importance of rationalizing the use of antibiotic in order to limit antibiotic resistance [3]. Antibiotics cause cell retardation and cell-death by targeting and inhibiting essential cellular processes [4]. If bacteria are exposed to drugs below the level required to kill bacteria in a population (MBC or Minimum Bactericidal Concentration) they can mutate and resist antibiotic treatment [5]. Mechanism involved in minimizing the effect of antibiotic by bacteria, is by acquiring or increasing expression of drug efflux pumps. These pumps are responsible for expelling drug from cytoplasm and their ability to access to their target [6]. Mutation is responsible for causing extensive changes in transcriptone and proteome of the bacterium and alters the several proteins responsible for impacting resistance [7]. As a result these mutations promote further evolution of bacteria in a multi-drug environment, in a drugspecific manner. Thus they both provide resistance and spur further development of resistance [8]. The two parameters involved in evolution of resistance are - Resistance variability and Dose sensitivity [9]. In 1882 Robert Koch, the German microbiologist discovered that TB is caused by MTB, which is rod-shaped and is responsible for infecting one-third of the total world’s population and death of the people [10]. There is an internationally recognized strategy known as DOTS (Direct Observation Treatment-short course) for delivering the basics TB case – finding and curing it. It is a management strategy for public health system that include political commitment, case-detection with quality assured bacteriology, chemotherapy for short period, ensuring patients’ adherence to treatment, adequate supply of drug and sound reporting and recording system [11]. Tuberculosis is caused by members of MTB – M. bovis, M. microti and M. africanum [12]. Drug resistant tuberculosis is defined as a case in which Tuberculosis excrete bacilli, resistant to one or more anti-tubercular drug [13]. There are generally three types of drug resistance – RRTB (Rifampicin Resistant TB), which occurs when rpoB gene in MTB gets mutated. This rpoB gene is responsible for encoding the β- sub unit of RNA polymerase [14]. MDR TB (Multi Drug Resistance TB, is resistant to Isoniazid and Rifampicin that are most effective first line TB drug [15]. XDR TB (Extensive Drug Resistance) is defined as the tuberculosis, which is resistant to Isoniazid+Rifampicin+at least 1 fluroquinolones+any of 2nd line injectable drugs-Kanamycin, Amikacin or Capreomycin [16]. Rifampicin is the first line drug used in treatment of Tuberculosis and its resistance is known as RRTB and by binding to beta-subunit of Mycobacterium tuberculosis, it inhibits Mycobacterial dependent RNA polymerase. rpoB gene is responsible for coding of beta subunit of RNA polymerase. Resistance to Rifampicin is indicated by mutation in rpoB gene [17]. Although acid fast bacilli (AFB) microscopy is most popular method for TB diagnosis but it has low sensitivity and is very slow method. In modern times Nucleic Acid Amplification (NAA) technology is being used due to their rapidity and sensitivity [18].
Materials and Methodology
The experiment was carried out at Central Molecular Research Laboratory, Department of Biochemistry, Shri Guru Ram Rai Institute of Medical and Health Sciences (SGRRIM&HS), Patel Nagar, Dehradun, Uttarakhand. Patients, who met the criteria of being TB positive cases, were recruited for the drug resistance study. Clinical specimens which includes Pulmonary such as Sputum, Pleural fluid, Bronchiolar alveolar lavage, bronchial secretions and extra pulmonary specimens such as Pus, Urine, Semen, Tissue, Endometrial blood, Cerebrospinal Fluid were considered for the study. Specimens were collected from patients attending Out Patient Department (OPDs) and In Patient Departments (IPDs) of Shri Mahant Indresh Hospital, Dehradun, (Uttarakhand) India. Collected samples were taken for further testing in areas:
Bio-safety cabinet: After collecting the sample, centrifugation process was done for isolating the DNA and the isolation was done by silica column method.
Pre-amplification area: This area was used for master mix preparation for PCR by targeting the specific gene (mpb64 and rpoB at 240 and 315 base pair respectively).
Amplification area: In this area the amplification or multiplication of targeted gene was done by PCR.
Post amplification area: After amplification the product was tested or analysed by using electrophoresis unit.
Results
The present study was undertaken to detect mycobacterial infection in pulmonary and extra- pulmonary samples collected from patients. We used PCR as a tool for detecting mycobacterial mpb64 gene and rpoB gene resistance in both pulmonary and extra-pulmonary samples collected from suspected TB patient. 70 samples were taken from the different Departments in Shri Mahant Indiresh Hospital and processed in Central Molecular Research Laboratory, Department of Biochemistry, Shri Guru Ram Rai Institutes of Medical and Health Sciences, Patel Nagar, Dehradun, Uttarakhand, India. On the basis of presence of mycobacterium, 2 samples were found positive in the process of AFB staining while 9 samples were found positive in the process of PCR for mpb64 gene (TB), from which 5 samples came positive for rpoB gene resistance.
Positivity rate for pulmonary and extra-pulmonary cases
Grouping the samples on the basis of site of infection, it can be divided into 2 sites, named as Pulmonary Site and Extra Pulmonary Site. Total clinical sample received were 70 and out these 60 i.e., 85.75% were from extra pulmonary site, while 10 i.e., 14.28% were from pulmonary site. For gene mpb64, TB PCR that was found to be positive was 3 (5%) and negative cases were 57 (95%). In case of rpoB gene, no PCR was found to be positive, while 3 samples gave 100% negative PCR. The total cases for pulmonary infection were 10 (14.28%) out of 70 obtained clinical samples. In case of pulmonary samples, PCR positive for mpb64 gene was 60% while PCR negative for it was 40%. The PCR positive for rpoB gene were 5 (83.33%) that means there was resistance towards Rifampicin due to mutation in rpoB gene while PCR negative for it was 1 (16.66%). These whole data indicates that pulmonary samples were found to be more positive in case of mpb64 and also the resistivity by rpoB gene was high in it as compared to extra pulmonary samples (Table 1; Figure 1).
| Total cases |
Positive cases of mpb64 gene |
Positive cases of rpoB gene |
| Extra-pulmonary site (60) |
3(5%) |
0(0%) |
| Pulmonary site (10) |
6(60%) |
5(83%) |
Table 1: Positive cases for mpb64 and rpoB gene in pulmonary and extra-pulmonary sites.
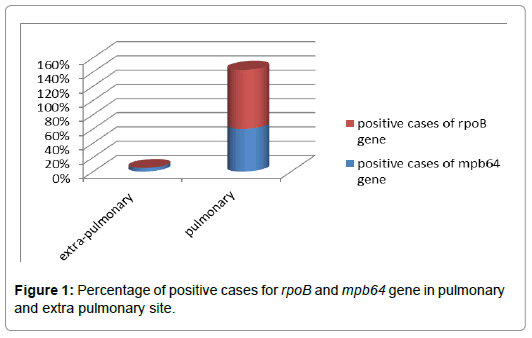
Figure 1: Percentage of positive cases for rpoB and mpb64 gene in pulmonary and extra pulmonary site.
Positivity and negativity rate for gender
Out of 70 received clinical samples; it was found that 30 samples were of females, which was 42.85% of total sample received and 40 were of males, which was 57.14%. In case of females, 3 samples (10%) were positive for mpb64 gene while 27 (90%) were negative for mpb64 gene but in case of rpoB gene 1 (33.33%) females showed positive response towards it, while 2 (66.66%) females showed negative response. In case of males, 6 (15%) were positive for mpb64 and 34 (85%) were negative towards it. While in case of rpoB gene, male 4 (66.66%) gave positive response by showing resistance to Rifampicin and 2 (33.33%) gave negative response by showing no mutation of rpoB gene. These whole data indicates that males are more prone to TB infection and also resistance towards Rifampicin in them is higher than females (Table 2; Figure 2).
| Gender |
Total Cases |
mpb64 gene |
rpoB gene |
| Positive |
Negative |
Positive |
Negative |
| Female |
30 (42.85%) |
3 (10%) |
27 (90%) |
1 (33.33%) |
2 (66.66%) |
| Male |
40 (57.14%) |
6 (15%) |
34 (85%) |
4 (66.66 %) |
2 (33.33%) |
Table 2: Positivity and negativity rate of mpb64 and rpoB gene in males and females.
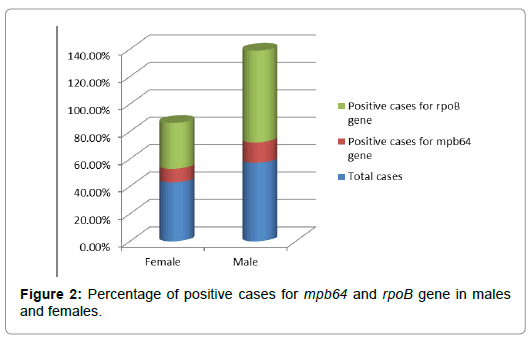
Figure 2: Percentage of positive cases for mpb64 and rpoB gene in males and females.
Clinical specimen wise result interpretation
The data give information regarding specimen type being used and its response towards mpb64 gene and rpoB gene. Out of the 70 specimen received, maximum sample was of CSF that was 38.5%. The BAL sample showed the maximum positive response that is 85%, towards mpb64 gene indicating them as TB positive. In case of rpoB gene also, BAL sample showed maximum positive response that is 83.33% making it highly resistant against Rifampicin (Table 3; Figure 3).
| |
Sample Types |
Cases |
mpb64 |
rpoB gene |
| Positive |
Negative |
Positive |
Negative |
| 1 |
Pus |
5 (7.14%) |
1 (20%) |
4 (80%) |
0 |
1 (20%) |
| 2 |
Tissue |
2 (2.85%) |
0 |
2 (2.85%) |
0 |
0 |
| 3 |
Endometrial blood |
2 (2.85%) |
0 |
2 (2.85%) |
0 |
0 |
| 4 |
Bronchio-alveolar lavage |
7 (10%) |
6 (85%) |
1 (14.28%) |
5 (83.33%) |
1 (16.66%) |
| 5 |
Cerebrospinal fluid |
27 (38.5%) |
1 (3.7%) |
26 (96.29%) |
0 |
1 (3.7%) |
| 6 |
Menstural blood |
1 (1.42%) |
0 |
1 (1.42%) |
0 |
0 |
| 7 |
Pleural fluid |
12 (17.14%) |
1 (8.33%) |
11 (91.66%) |
0 |
1 (8.33%) |
| 8 |
Ascitic fluid |
7 (10%) |
0 |
7 (10%) |
0 |
0 |
| 9 |
Pericardial fluid |
1 (1.42%) |
0 |
1 (1.42%) |
0 |
0 |
| 10 |
Sputum |
2 (2.85%) |
0 |
2 (2.85%) |
0 |
0 |
| 11 |
Urine |
2 (2.85%) |
0 |
2 (2.85%) |
0 |
0 |
| 12 |
Synovial fluid |
1 (1.42%) |
0 |
1 (1.42%) |
0 |
0 |
Table 3: Percentage of available sample types and their response.
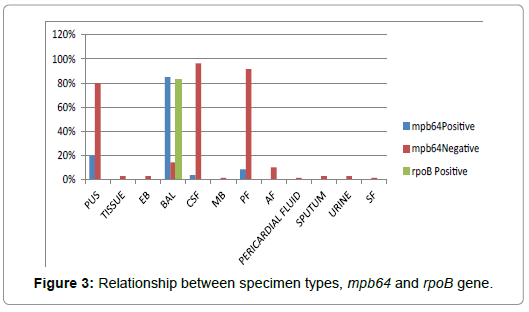
Figure 3: Relationship between specimen types, mpb64 and rpoB gene.
Discussion
Global prevalence of infection due to MTB is 32% and enormity of the problem has increased with emergence of Rifampicin resistant strain of Mycobacterium tuberculosis. Patient at high risk of MDR TB are need to be screened, so for this purpose rapid, inexpensive and simple techniques are used. One technique can be mentioned to the rapid molecular methods, is based on amplification of DNA (NAA), which is used in order to follow the mutation in genes that are responsible for resistance to Rifampicin (rpoB gene) and other anti-tubercular drugs. After working on 70 samples during our lab scale work we concluded and analyzed the presence of mycobacterium in various samples of patients. Out of these 70 samples, 9 were found to be TB PCR positive for mpb64 gene and out of these 9 positive samples, 5 samples showed positive response for rpoB gene resistance, that means mutation in rpoB gene of Mycobacterium tuberculosis leading in resistance to Rifampicin drug. On the basis of gender we concluded that male were found to be more TB infected than female as indicated from table above. Percentage of males those were infected to mycobacterium was 15%, while females were 10%. In case of rpoB gene resistance also, males were higher in percentage and were found to be 66.66% and females were with percentage of 33.33%. In case of ‘Site of Infection’ by mycobacterium, extra-pulmonary site was found to be more infected than pulmonary site. In case of pulmonary TB infection, BAL sample showed most TB positive cases. 60% patients showed PCR positive for mpb64 gene and 83.33% showed resistance to rpoB gene. While in case of extra pulmonary infection, positive response given by mpb64 gene was only 5% and rpoB gene gave no response. By analyzing the result of specimen being used and its response towards mpb64 gene and rpoB gene, we concluded that BAL sample gave 85% positive response for mpb64 and 83.33% gave positive response for rpoB gene. Second highest positive response for mpb64 was given by Pus sample but it gave no positive response towards rpoB gene (Figure 4).
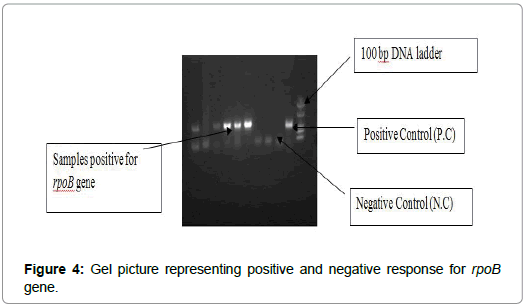
Figure 4: Gel picture representing positive and negative response for rpoB gene.
Conclusion
Resistance towards any anti-tubercular drug, act as an obstacle in the treatment of tuberculosis. The cause for drug resistance of Rifampicin could be patient’s incompliance with their prescription as well as easy availability of drug (Rifampicin) at the drug counter for nose and throat infection. Carelessness in the diagnosis of TB may lead to drug resistance, which can affect the duration of TB treatment. mpb64 gene is not sufficient for the diagnosis of drug resistance in TB, so for this purpose rpoB gene (responsible for drug resistance) is targeted.
Acknowledgments
The authors are grateful to Honorable Chairman, Shri Guru Ram Rai Education Mission and faculties of Uttarakhand Technical University for their kind support and guidance.
24362
References
- Levy SB, Marshall B (2004) Antibacterial resistance worldwide: Causes, challenges and responses. Nature Med 10: S122.
- Wirsing RL, Logan MH, Micozzi MS, Nyamwaya DO, Pearce TO, et al. (1985) The health of traditional societies and the effects of acculturation [and comments and replies]. Curr Anthropol 26: 303-322.
- O’brien DJ, Gould IM (2013) Maximizing the impact of antimicrobial stewardship: the role of diagnostics, national and international efforts. Current Opinion in Infectious Diseases 26: 352-358.
- Zhang H, Burrows F (2004) Targeting multiple signal transduction pathways through inhibition of Hsp90. J Mol Med 82: 488-499.
- Coates A, Hu Y, Bax R (2002) The future challenges facing the development of new antimicrobial drugs. Nature Rev Drug Discovr 11: 895.
- Tenover FC (2006) Mechanisms of antimicrobial resistance in bacteria. American J Infect Conf 34: S3-S10.
- Vranakis I, Goniotakis I, Psaroulaki A, Sandalakis V, Tselentis Y, et al. (2104) Proteome studies of bacterial antibiotic resistance mechanisms. J Proteomics 97: 88-99.
- Richardson LA (2017) Understanding and overcoming antibiotic resistance. PLoS Biol 15: e2003775.
- Kahn SE (2003) The relative contributions of insulin resistance and beta-cell dysfunction to the pathophysiology of type 2 diabetes. Diabetologia 46: 03-19.
- Godreuil S, Tazi L, Bañuls AL (2007) Pulmonary tuberculosis and Mycobacterium Tuberculosis: modern molecular epidemiology and perspectives. Encyclopaedia of Infectious Diseases: Modern Methodologies. Hoboken, NJ: John Wiley & Sons.
- World Health Organization (2015) Implementing the end TB strategy: The essentials.
- Cosma CL, Sherman DR, Ramakrishnan L (2003) The secret lives of the pathogenic mycobacteria. Ann Rev Microbiol 57: 641-676.
- Telenti A (1997) Genetics of drug resistance in tuberculosis. Clin Chest Med 18: 55-64.
- Mehta B, Siddiquie A, Kaushik R, Bisht R, Sharma N (2015) Amplification of rpob, kat g and mab a (fab g1)-inh a promotor DNA sequences by PCR in multiple drug resistance tuberculosis. Int J Life Sci Sci Res 1: 15-18.
- Sharma N, Sachan SK, Bushan I, Rai R, Shukla A, et al. (2017) Assessment of extensively drug resistant tuberculosis by studying the drug sensitivity pattern in north Indian region. J Microbiol Biotechnol Res 4: 31-33.
- Xu J, Tozawa Y, Lai C, Hayashi H, Ochi K (2002) A Rifampicin resistance mutation in the rpoB gene confers ppGpp-independent antibiotic production in Streptomyces coelicolor A3 (2). Molecular Genetics and Genomics 268: 179-189.
- Narotam S, Veena S, Raj SP, Kushwaha RS, Shivani S (2012) Conventional PCR usage for the detection of Mycobacterium Tuberculosis complex in Cerebrospinal Fluid by MPB64-Target PCR. Int J Drug Dev Res 4: 206-210.










