Parimal Chandra Bhomick1, Salam Pradeep Singh2*,Chitta Ranjan Deb3, Dipak Sinha1, Lakshmi Narayan Kakati4 and Bolin Kumar Konwar5
1Department of Chemistry, Nagaland
University, Lumami-798627, Nagaland,
India
2Bioinformatics Infrastructure Facility,
Nagaland University, Lumami-798627,
Nagaland, India
3Department of Botany, Nagaland
University, Lumami-798627, Nagaland,
India
4Department of Zoology, Nagaland
University, Lumami-798627, Nagaland,
India
5Department of Molecular Biology
and Biotechnology, Tezpur University,
Tezpur-784028, Assam, India (On-lien
Vice Chancellor, Nagaland University,
Lumami-798627, Nagaland, India)
- *Corresponding Author:
- Salam Pradeep Singh
Bioinformatics Infrastructure Facility,
Nagaland University, Lumami-798627,
Nagaland, India.
Tel: +918486268935
E-mail: salampradeep@gmail.com
Received date: February 19, 2016; Accepted date: May 05, 2016; Published date: May 11, 2016
Citation: Bhomick PC, Singh SP, Deb CR,
et al. Molecular Interaction Studies of
Chitosan Cross-linked Compounds as Drug
Delivery Substrate for Anticancer Agents.
J Biomedical Sci. 2016, 5:3. doi:10.4172/2254-609X.100032
Copyright: © 2016 Bhomick PC. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Introduction
Hydrogels are cross-linked polymeric networks that trap water in the polymer matrices mainly by surface tension and are usually three dimensional while most of its properties can be altered by structural modification [1,2]. A kind of hydrogel called chitosan is finding wide application in many areas such as fuel cells, oil encapsulation, wound dressing and especially in drug delivery in recent years [3-6]. Chitosan [β-(1,4)-2-amino-2-deoxy-dglucopyranose] is a non-toxic biodegradable hetero polymer, has a good absorption and adhesion property. It is a weak base obtained by deacetylation of chitin [3]. Because of their affinity for water and high degree of solubility in acidic medium, chitosan hydrogels comparatively shows low mechanical strength and minimum ability to control the delivery of encapsulated compounds [7], thereby facilitating chemical modification by its amino and hydroxyl groups. For a hydrogel to be introduced in a biological systems, its compatibility should be taken into account while it is seen that the pharmacy world will benefit from hydrogels like chitosan because of its hydrophilicity, flexibility, versatility, high water absorptivity, and greater compatibility with the biological system. It is also a choice for the pharmaceutical world because of its long life span in circulation and possibility of being actively or passively targeted to the known biophase like cancer cells [8]. Chitosan can be blended with different cross linking agent to produce a chemically active hydrogels for bioapplications. Like the amino group of chitosan can lead to ionic interaction between and anionic groups. These interactions can produce hydrogels with different material properties which depend upon the size of the anionic agents and charge density, also on degree of deacetylation and amount of chitosan polymer.
Chitosan cross linked with polyelectrolytes have been produced by water-soluble negatively charged molecules like DNA, alginate, hyaluronic acid, proteins like gelatin polyacrylic acid and mostly the stability of these compounds depends on solvent, charge density, temperature, ionic strength, and pH [9-12].
The NOS enzymes consist of oxygenase domain that binds arginine, tetrahydrobiopterin and heme and reductase domain with FAD and FMN prosthetic groups. They are complex, homodimeric heme enzyme which produces free radical nitric oxide that leads to variety of age-related diseases [13-15]. Nitric oxide synthase (NOS) in cells are of three isoforms. Neuronal/brain NOS (nNOS), endothelial NOS (eNOS) and inducible NOS (iNOS). nNOS and eNOS belongs to constitutive NOS (cNOS) and NO produced from this type helps in maintaining normal vasoactivity through a Ca2+ -dependent pathway and also as a neurotransmitter for signal transmission. While NO produced from iNOS can trigger several disadvantage cellular responses and can cause some diseases including sepsis, stroke and inflammation, [13-19].
In the present investigation chitosan cross linked compounds were screened as inhibitors of iNOS revealing chitosan as a good carrier for delivering and unloading the drug at specific target.
Materials and Method
Chemical dataset
The 2D structure of Diethylsquarate, Glutaraldehyde, Formaldehyde, Ethyleneglycol diglycildylether, Blocked Diisocyanate, Phloretic and Activated Quinone cross-linked with Chitosan was generated with Chemoffice 2010. The energy of these cross-linked compounds were further optimized using MM2 force field method and save as sybyl mol2 (three dimensional) file format using ChemOffice 2010.
Protein preparation
The 3D structure of human inducible nitric oxide synthase (PDB ID: 4NOS) was downloaded from the Protein Databank Bank (https://www.rcsb.org/). The coordinates of this enzyme is complexed with water molecules and iron protoporphyrin IX (heme) along with the Cofactors such as BH4, Zn+2 atom. Moreover the 3D structure has a resolution of 2.25 Å. making it an excellent choice for molecular docking studies. All the water molecules were removed for the molecular docking simulation purpose since they are not taken into account during the scoring function [20].
Docking computation
The 3D structure of human inducible nitric oxide synthase (PDB ID: 4NOS) was then imported in Molegro Virtual Docker (MVD). The sidechains conformations of 4NOS were further minimized using PLP-potentials for steric and hydrogen bonding interactions, and the Coulomb potential for the electrostatic forces. And only the torsion angles are modified during the minimization which includes bond lengths and backbone atom positions are held fixed [20].
Further, the potential ligand binding site of the enzyme was predicted using MVD. The binding site have a volume of 470.02 A3 and 1158.84 A2. The binding site was set inside a restriction sphere of radius 15 A° (X 0.65, Y 99.58, Z 11.19) using MVD.
Then the Chitosan cross linked compounds were then imported in MVD. The Bond flexibility of the cross likned compounds was set as well as the side chain flexibility of residues near the potential ligand binding site was set with a tolerance of 1.10 and strength of 0.90 for docking simulations. The RMSD threshold was set at 2.00 Å for multiple cluster poses. The docking algorithm was set at 1,500 maximum iteration with simplex evolution size of 50 and a minimum of 20 runs were performed for each of the cross linked compound. The best pose was considered for subsequent protein-ligand interaction analysis.
Molecular docking was carried out using MVD which is based on a differential evolution algorithm. The algorithm of MVD considers the sum of the intermolecular interaction energy between the ligand and the protein and the intramolecular interaction energy of the ligand. The docking energy scoring function is based on the modified piecewise linear potential (PLP) with new hydrogen bonding and electrostatic terms included. Full description of the algorithm and its reliability compared to other common docking algorithm is described by Thomsen et al. [20].
Results and Discussion
The molecular docking simulation revealed the cross linked compounds bind at the active site of the NOS. From the docking score, it is revealed that the MolDock score holds favorable interaction for the Chitosan Cross linked compounds viz. Diethyl Squarate (-181.13 kJmol-1), Formaldehyde (-179.522 kJmol-1), Glutaraldehyde (-145.48 kJmol-1), Blocked Diisocynate (-105.27 kJmol-1), Activated Quinone (-104.72 kJmol-1) in terms of negative energy (Table 1). While Ethyleneglycol diglycildyl ether, Phloretic acid and Genepindo not possessed a favourable interaction.
| Chitosan Cross Linked Ligands |
MolDock Score |
Interaction |
HBond |
LE1 |
LE3 |
| Diethyl Squarate |
-181.13 |
-313.47 |
-7.75 |
-1.46 |
0.17 |
| Formaldehyde |
-179.52 |
-259.00 |
-14.09 |
-1.51 |
2.25 |
| Glutaraldehyde |
-145.48 |
-245.14 |
-12.19 |
-1.22 |
2.12 |
| Blocked Diisocyanate |
-105.27 |
-210.05 |
-10.23 |
-0.81 |
1.82 |
| Activated Quinone |
-104.73 |
-207.40 |
-8.29 |
-0.84 |
3.18 |
| Ethyleneglycoldiglycildyl ether |
-35.45 |
-105.79 |
-8.16 |
-0.27 |
2.94 |
| Phloretic acid |
24.85 |
-124.02 |
-10.23 |
0.19 |
2.25 |
| Genepin |
90.39 |
20.50 |
-3.59 |
0.61 |
9.12 |
Table 1: Molecular docking score of the Chitosan cross linked compounds
The MolDock scoring function (MolDock Score) which determined the binding affinity in the present investigation is derived from the Piecewise Least Potential (PLP) scoring functions. The MolDock scoring function further improves the PLP scoring functions with new hydrogen bonding term and new charge schemes. The docking scoring function, EScore, is defined by the following energy terms:
EScore = Einter + Eintra
Where,
Einter is the ligand-protein interaction energy
Eintra is the internal energy of the ligand
Further, the ligand-protein interaction calculation using ligand energy inspector for the Chitosan cross linked compounds is shown in Table 2 which indicates the ligand-protein interaction energy and its molecular interaction distances along with the interacting atoms. It is observed that the compounds also exhibit molecular interaction with the Cofactor HEM molecule. Thus, indicating a strong binding affinity towards the active site of NOS. The snap shots illustrating the protein-ligand interaction of Chitosan cross linked with Diethyl Squarate, Formaldehyde, Glutaraldehyde, Blocked Diisocyanate and Activated Quinone is shown in Figures 1-5 respectively. Hence, from the figures the plausibility of Chitosan cross linked compounds delivering ligands at a given specific target is viable.
| Chitosan Cross Linked Ligands |
Ligand--- Protein Interaction |
Interaction energy |
Interaction Distance |
Diethyl squarate
 |
N(54)---Glu494(OE1) |
-2.5 |
2.87 Å |
| O(97)---Arg388(NH2) |
-0.28 |
2.73 Å |
| O(65)---Gln263(NE2) |
-2.5 |
2.90 Å |
| O(101)---Asn283(OD1) |
-2.44 |
2.90 Å |
| O(106)---Ala282(O) |
-1.82 |
3.23 Å |
| O(28)---Val386(N) |
-1.88 |
2.85 Å |
| O(26)---Thr121(OG1) |
-0.59 |
3.48 Å |
| O(36)---Asp385(OD2) |
-2.02 |
3.20 Å |
| O(95)---Tyr373(OH) |
-0.67 |
3.47 Å |
| O(101)---Agr266(NH1) |
-2.5 |
2.93 Å |
| O( 77) ---Agr266(NH1) |
-0.38 |
3.52 Å |
| N(112)--- Ala282(O) |
-0.65 |
2.82 Å |
| O(70)--- Gln263(NE2) |
-0.19 |
3.56 Å |
| O(68)--- Gln263(NE2) |
-0.01 |
3.60 Å |
| O(97)---Asp383(OD1) |
-1.78 |
2.51 Å |
| O(93)---Trp372(O) |
-0.96 |
2.42 Å |
| O(93)---Glu377(OE2) |
-0.92 |
2.41 Å |
| O(103)---Glu377(OE1) |
-1.46 |
3.31 Å |
| O(93)---Met374(N) |
-0.60 |
3.40 Å |
| O(102)---N(16) HEM Cofactor |
-1.26 |
3.35 Å |
| O(102)---N(24) HEM Cofactor |
-2.19 |
3.16 Å |
| N(107)---N(24) HEM Cofactor |
-1.60 |
3.28 Å |
| N(107)---N(32) HEM Cofactor |
-2.5 |
3.01 Å |
| N(108)---O(14) HEM Cofactor |
-1.16 |
3.37 Å |
| O(103) ---O(14) HEM Cofactor |
-0.93 |
3.41 Å |
Table 2a: Molecular interaction analysis of Chitosan cross linked Diethyl squarate
Formaldehyde
 |
O(93)---Lys497(NZ) |
-2.5 |
3.06 Å |
| O(93)---Glu494(OE1) |
-2.5 |
2.98 Å |
| O(51)---Gln492(N) |
-1.35 |
3.33 Å |
| O(34)---Glu285(OE1) |
-2.02 |
3.20 Å |
| O(95)---Thr121(OG1) |
-2.5 |
2.92 Å |
| O(87)--- Thr121(OG1) |
-0.56 |
3.49 Å |
| N(54)---Pro350(O) |
-2.5 |
2.96 Å |
| O(47)--- Phe369(O) |
-0.82 |
3.44 Å |
| O(40)---Ala262(O) |
-2.5 |
2.73 Å |
| O(38)--- Ala262(O) |
-1.24 |
3.35 Å |
| O(38)---Asn354(ND2) |
-2.5 |
2.73 Å |
| O(97)---Asp385(OD2) |
-1.74 |
3.25 Å |
| O(70)---Arg381(NH2) |
-0.96 |
3.39 Å |
| O(72)--- Arg381(NH2) |
-0.57 |
3.45 Å |
| O(72)--- Arg381(NH1) |
-1.24 |
3.35 Å |
| O(105)--- Arg381(NH1) |
-0.67 |
2.56 Å |
| O(42)---Glu377(OE1) |
-2.5 |
3.08 Å |
| O(101)---IIe119(O) |
-2.38 |
3.00 Å |
| O(56)---Tyr373(OH) |
-0.69 |
2.78 Å |
| O(101)--- Arg199(NH2) |
-0.17 |
3.36 Å |
| O(45)---O(41) HEM Cofactor |
-0.48 |
3.50 Å |
| N(52)---O(14) HEM Cofactor |
-1.08 |
3.38 Å |
| N(111)---O(14) HEM Cofactor |
-1.45 |
3.31 Å |
Table 2b: Molecular interaction analysis of Chitosan cross linked Formaldehyde
Glutaraldehyde
 |
O(46)---Arg381(NH1) |
-2.5 |
2.92 Å |
| O(46)--- Arg381(NH2) |
-1.61 |
2.61 Å |
| O(110)---Tyr347(OH) |
-2.5 |
2.68 Å |
| O(65)---Gln263(NE2) |
-2.5 |
2.74 Å |
| O(101)---Glu494(OE1) |
-0.63 |
3.47 Å |
| H(180)---Gln492(O) |
-2.5 |
1.76 Å |
| N(113)---Asn354(O) |
-2.5 |
2.75 Å |
| O(106)---Asn354(OD1) |
-2.5 |
2.94 Å |
| O(99)---Tyr491(OH) |
-1.95 |
3.21 Å |
| O(56)---Trp463(NE1) |
-0.38 |
2.94 Å |
| O(42)---Met120(N) |
-0.52 |
3.05 Å |
| O(102)---Trp372(O) |
-2.5 |
2.92 Å |
| N(107)---Glu377(OE1) |
-0.07 |
2.31 Å |
| O(95)---O(41) HEM Cofactor |
-2.5 |
2.82 Å |
| O(102)---N(16) HEM Cofactor |
-2.5 |
3.09 Å |
| O(99)---O(42) HEM Cofactor |
-0.20 |
3.56 Å |
| H(177)---O(41) HEM Cofactor |
-2.5 |
1.82 Å |
Table 2c: Molecular interaction analysis of Chitosan cross linked Glutaraldehyde
Blocked diisocyanate
 |
N(113)---Thr121(O) |
-2.5 |
2.99 Å |
| O(106)---Thr121(N) |
-1.99 |
3.20 Å |
| O(36)---Glu494I(OE2) |
-2.32 |
2.58 Å |
| H(152)---Ala282(O) |
-2.5 |
1.95 Å |
| O(45)---Thr121(OG1) |
-0.74 |
3.45 Å |
| N(52)--- Thr121(OG1) |
-0.02 |
2.30 Å |
| O(127)---Arg266(NH1) |
-2.01 |
2.83 Å |
| N(48)---Ala282(O) |
-0.95 |
3.12 Å |
| O(34)---Trp496(NE1) |
-0.68 |
2.64 Å |
| O(44)---Asn354(ND2) |
-2.5 |
3.09 Å |
| O(129)---Arg381(NH2) |
-1.97 |
2.66 Å |
| O(87)--- Arg381(NH1) |
-2.5 |
2.64 Å |
| O(79)--- Arg381(NH1) |
-2.28 |
3.14 Å |
| O(79)--- Arg381(NH2) |
-0.15 |
3.56 Å |
| O(102)---O(15) HEM Cofactor |
-2.5 |
2.61 Å |
| O(102)---O(14) HEM Cofactor |
-1.23 |
2.45 Å |
| O(46)---O(41) HEM Cofactor |
-1.28 |
3.35 Å |
| N(53)---O(41) HEM Cofactor |
-1.88 |
3.22 Å |
Table 2d: Molecular interaction analysis of Chitosan cross linked Blocked diisocyanate
Activated Quinone
 |
N(107)---Ser118(O) |
-2.45 |
3.11 Å |
| O(93)---Arg381(NH1) |
-2.5 |
2.91 Å |
| O(65)--- Arg381(NH1) |
-2.5 |
2.60 Å |
| O(63)--- Arg381(NH1) |
-0.08 |
3.58 Å |
| O(65)--- Arg381(NH2) |
-0.22 |
3.31 Å |
| O(63)--- Arg381(NH2) |
-0.03 |
3.59 Å |
| O(43)---Arg388(NH2) |
-0.65 |
3.20 Å |
| O(43)---Asp382(OD1) |
-1.84 |
3.23 Å |
| O(43)---Gln263(NE2) |
-0.12 |
3.56 Å |
| O(45)--- Gln263(NE2) |
-2.37 |
3.13 Å |
| O(26)---Asn354(ND2) |
-0.97 |
3.41 Å |
| O(18)--- Asn354(ND2) |
-1.57 |
3.29 Å |
| O(42)--- Asn354(ND2) |
-2.06 |
3.07 Å |
| O(42)---Asn354(O) |
-1.48 |
2.48 Å |
| O(40)--- Asn354(OD1) |
-2.37 |
2.58 Å |
| O(47)---Thr121(OG1) |
-2.5 |
2.90 Å |
| N(54)---Glu494(OE1) |
-2.5 |
2.88 Å |
| O(28)---Tyr373(OH) |
-1.14 |
3.37 Å |
| O(20)---Tyr347(OH) |
-2.5 |
2.97 Å |
| N(48)--- Tyr347(OH) |
-2.5 |
2.84 Å |
| O(36)--- Tyr347(OH) |
-2.5 |
2.94 Å |
| O(43)---Arg388(NH1) |
-2.5 |
2.96 Å |
| O(79)---Trp463(NE1) |
-0.15 |
2.95 Å |
| O(44)---Tyr373(OH) |
-0.35 |
3.32 Å |
| O(44)---Glu377(OE1) |
-1.89 |
2.53 Å |
| N(49)---Glu377(OE1) |
-1.27 |
3.16 Å |
| O(34)---O(15) HEM Cofactor |
-1.48 |
3.30 Å |
| N(108)---O(15) HEM Cofactor |
-1.86 |
2.52 Å |
| N(108)---O(14) HEM Cofactor |
-0.97 |
3.41 Å |
| O(104)---O(14) HEM Cofactor |
-1.77 |
3.25 Å |
| O(34)---O(14) HEM Cofactor |
-0.84 |
3.43 Å |
| O(46)---O(41) HEM Cofactor |
-0.07 |
3.59 Å |
Table 2e: Molecular interaction analysis of Chitosan cross linked Activated Quinone
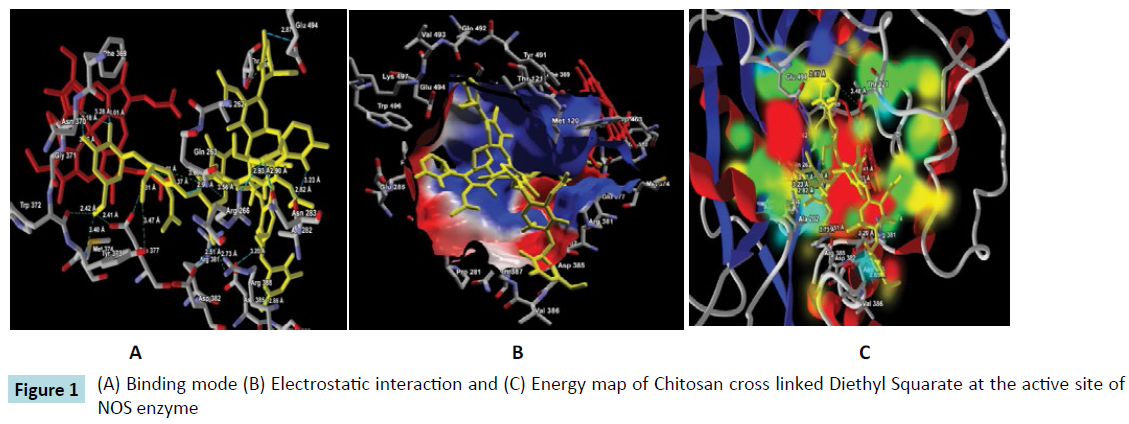
Figure 1: (A) Binding mode (B) Electrostatic interaction and (C) Energy map of Chitosan cross linked Diethyl Squarate at the active site of NOS enzyme
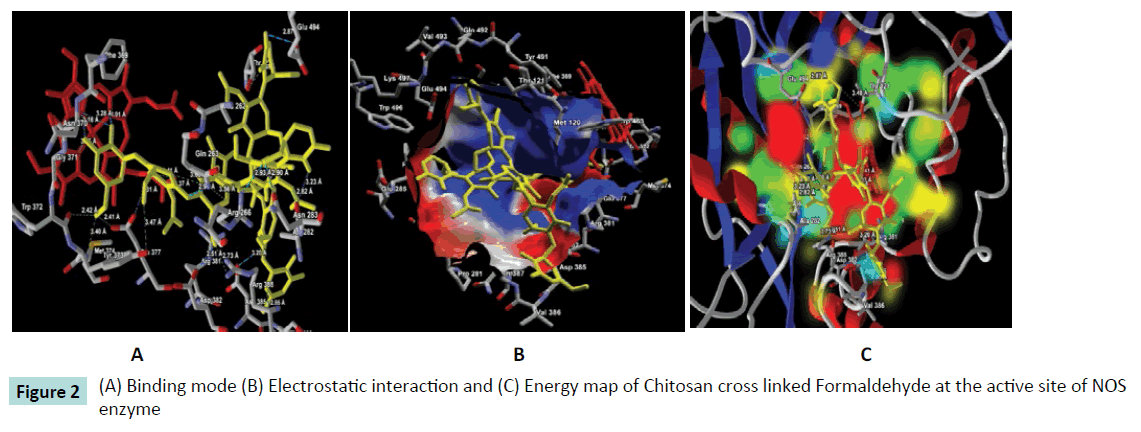
Figure 2: (A) Binding mode (B) Electrostatic interaction and (C) Energy map of Chitosan cross linked Formaldehyde at the active site of NOS enzyme
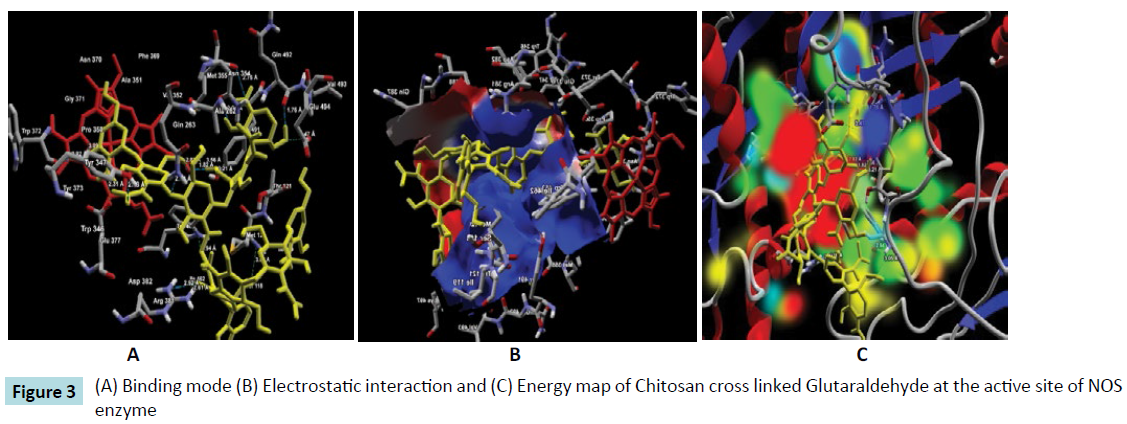
Figure 3: HPLC chromatogram of the nine reference compounds in 50% aqueous methanol, measured at 370nm. Retention times for rutin, sutherlandin A, sutherlandin B, kaempferol-3-O-rutinoside, sutherlandin C, sutherlandin D, quercitrin, quercetin and kaempferol were 11.9, 12.7, 13.8, 15.3, 16.2, 17.0, 18.0, 26.2 and 28.1 minutes, respectively.
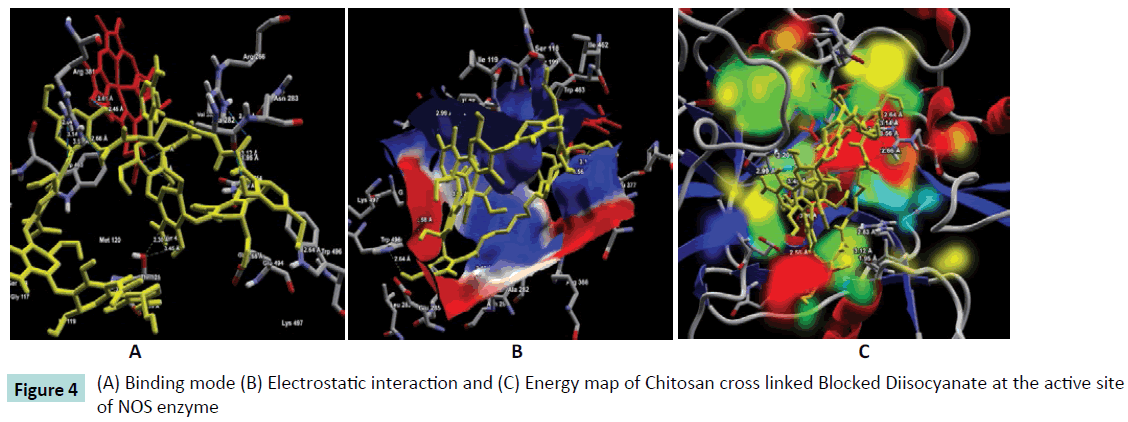
Figure 4: (A) Binding mode (B) Electrostatic interaction and (C) Energy map of Chitosan cross linked Blocked Diisocyanate at the active site of NOS enzyme
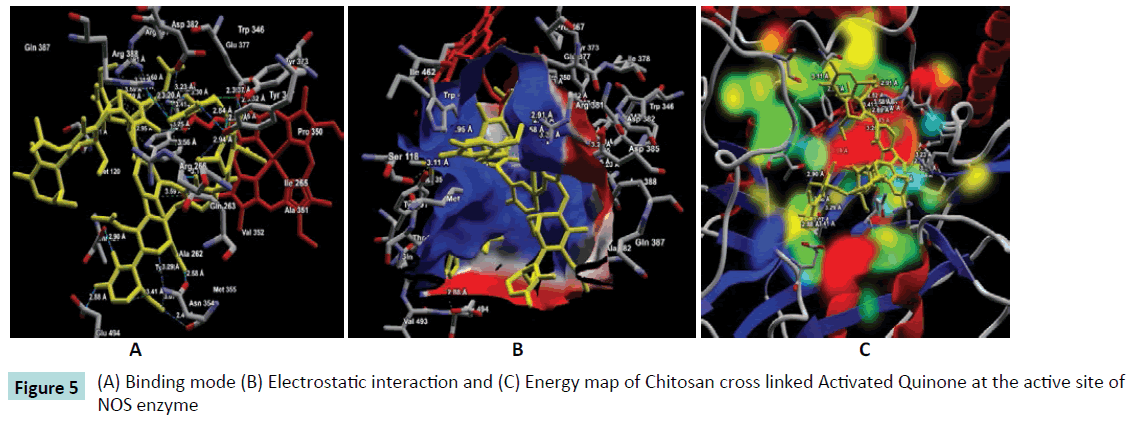
Figure 5: (A) Binding mode (B) Electrostatic interaction and (C) Energy map of Chitosan cross linked Activated Quinone at the active site of NOS enzyme
Conclusion
To conclude, molecular docking simulation was carried out against NOS enzyme with a set of Chitosan cross linked compounds. The molecular docking results showed favourable binding affinity and docking score of the majority of the compounds at the active site of NOS enzyme. The molecular interaction analysis also revealed heavy molecular interaction with the active site residues. Thus, we bring to a cause that Chitosan cross linked compounds is a good inhibitor of NOS enzyme as anticancer agents. Moreover, Chitosan is successful in delivering the compounds at the specific active site of the target enzyme.
Conflict of interest
The authors declare no conflict of interest exist
Acknowledgement
The authors would like to acknowledge the Department of Biotechnology, Ministry of Science and Technology, Government of India, New Delhi for the necessary support. PCB acknowledged the Department of Science and Technology (DST), Government of India for DST-INSPIRE fellowship (IF150297).
9390
References
- Choudhury NA, Sampath S, Shukla AK (2009) Hydrogel-polymer electrolytes for electrochemical capacitors: an overview. Energy Environ Sci 2: 55-67
- Giri TK, Thakur A, Alexander A (2012) Modified chitosan hydrogels as drug delivery and tissue engineering systems: present status and applications. ActaPharmaceuticaSinica B 2: 439-449.
- Ma J, Choudhury NA, Sahai, et al. (2011) A high performance direct borohydride fuel cell employing cross-linked chitosan membrane. Power Sources 196: 8257-8264.
- Bhattarai N, Gunn J, Zhang M (2010) Chitosan-based hydrogels for controlled, localized drug delivery. Adv Drug Deliv Rev 62: 83-99.
- Abreu FO, Oliveira EF, Paula HC, de Paula RC (2012) Chitosan/cashew gum nanogels for essential oil encapsulation. CarbohydrPolym 89: 1277-1282.
- MogoÅŸanu GD, Grumezescu AM (2014) Natural and synthetic polymers for wounds and burns dressing. Int J Pharm 463: 127-136.
- Crinia G, Badot PM (2008) Application of chitosan, a natural aminopolysaccharide, for dye removal from aqueous solutions by adsorption processes using batch studies: A review of recent literature. ProgPolymSci 33: 399-447.
- Hamidi M, Azadi A, Rafiei P (2008) Hydrogel nanoparticles in drug delivery. Adv Drug Deliv Rev 60: 1638-1649.
- Brack HP, Tirmizi SA, Risen WM (1997) A spectroscopic and viscometric study of the metal ion-induced gelation of the biopolymer chitosan. Polymer 38: 2351-2362.
- Dambies L, Vincent T, Domard A, Guibal E (2001) Preparation of chitosan gel beads by ionotropicmolybdate gelation. Biomacromolecules 2: 1198-1205.
- Singh SP, Konwar BK (2012) Molecular docking studies of quercetin and its analogues against human inducible nitric oxide synthase. Springerplus 1: 69.
- Tsuchida E, Abe K (1982) Interactions between Macromolecules in Solution and Intermacromolecular Complexes. AdvPolymSci 45: 1-119.
- Garthwaite J, Boulton CL (1995) Nitric oxide signaling in the central nervous system. Annu Rev Physiol 57: 683-706.
- Stuehr DJ (1997) Structure-function aspects in the nitric oxide synthases. Annu Rev PharmacolToxicol 37: 339-359.
- Chung HY, Cesari M, Anton S, Marzetti E, Giovannini S, et al. (2009) Molecular inflammation: underpinnings of aging and age-related diseases. Ageing Res Rev 8: 18-30.
- Marletta MA, Yoon PS, Iyengar R, Leaf CD, Wishnok JS (1988) Macrophage oxidation of L-arginine to nitrite and nitrate: nitric oxide is an intermediate. Biochemistry 27: 8706-8711.
- Nathan C (1992) Nitric oxide as a secretory product of mammalian cells. FASEB J 6: 3051-3064.
- Yen-Chou C, Shing-Chuan S, Woan-Ruoh L, Wen-Chi H, et al. (2001) Inhibition of nitric oxide synthase inhibitors and lipopolysaccharide induced inducible NOS and cyclooxygenase-2 gene expressions by rutin, quercetin, and quercetinpentaacetate in RAW 264.7 macrophages. J Cell Biochem 82: 537-548.
- Fischmann TO, Hruza A, Niu XD, Fossetta JD, Lunn CA, et al. (1999) Structural characterization of nitric oxide synthase isoforms reveals striking active-site conservation. Nat StructBiol 6: 233-242.
- Thomsen R, Christensen MH (2006) MolDock: a new technique for high-accuracy molecular docking. J Med Chem 49: 3315-3321.