Keywords
STEC, non-STEC, Multidrug resistance, PCR, tetA,tetB and tetC, strA and strB.
Introduction
Escherichia coli is a common inhabitant of the intestinal tract of a wide variety of animals and humans [1] and this intestinal bacteria can be easily disseminated in different ecosystems through the water, soil, food and others [2]. The increase in antimicrobial resistance and the impact on human health is an emerging problem worldwide. Exposure to resistant bacteria through food chain has gained increased attention, as the presence of resistant bacteria in food and water might have an impact on the development and dissemination of resistance to antimicrobial agents in human bacterial pathogens. A recent study conducted by Johnson et al [3] suggests that retail foods, especially meat and meat products, may be an important vehicle for community-wide dissemination of antimicrobial resistant Escherichia coli and extraintestinal pathogenic Escherichia coli. Antibiotic usage is probably the most important factor that promotes the emergence, selection and dissemination of antibiotic-resistant in both veterinary and human medicine [4; 5]. In addition to their therapeutic use in human and veterinary medicine, antimicrobials are routinely used for disease prevention and growth promotion agents in animal production. It has been shown that repeated sublethal exposure to antimicrobial agents not only promotes adaptive resistance but also confers decreased sensitivity to antibiotics [6].
The aim of the present study was to assess the actual frequency of antimicrobial resistance in shiga toxin-producing Escherichia coli (STEC) and non-shiga toxin-producing E. coli from various meats and ready-to-eat (RTE) meat products, water and human sources in Punjab, India at the phenotype level. The second objective was to assess the diversity and distribution of the major resistance genes to tetracyclines and streptomycin in these STEC isolates and other non-STEC isolates from the above sources.
Materials and methods
Sample collection
A total of 675 samples comprising 392 raw meats (pork n=106, buffalo meat n= 69, chevon n=68, mutton n=55, fish n=54 and chicken=40), 167 RTE meat products (buffalo meat n=40, chicken n=40, fish n=27, pork n=25, mutton n=25 and chevon n=10) from different retail meat shops in Punjab, drinking water samples (n=64) from different localities in Punjab and human diarrhoeic samples (n=52) from different Government and corporate hospitals in Punjab were collected between June 2008 and December 2009.Approximately 50g of the raw meat and RTE meat products, 500 ml of drinking water samples and 10g of human diarrhoeic samples were collected aseptically in sterile wide mouth screw capped disposable polyethylene containers and transported to the laboratory on ice and processed the sample for isolation of E. coli with in 24h of sample collection
Bacterial isolates
Two hundred fifty three E. coli isolates were isolated from different raw meats (n=192), RTE meat foods (n=28), drinking water (n=19) and human diarrhoeic samples (Children below 10 years) (n=14) by standard procedure. The samples were enriched in Tryptic soy broth (TSB), then streaked on EMB and MeConkey lactose (MLA)agar plates. Isolates showing greenish metallic sheen growth on EMB and pink colour colonies with central depression on MLA were picked up (single colony) and streaked on Nutrient agar slants and stored at 40C for further biochemical studies and for antimicrobial susceptibility tests. The identification of each isolate was confirmed by using the following tests: lack of oxidase activity, catalase, indole, methyl red (MR) and nitrate reduction positivity, voges proskauer (VP), citrate, urease, H2S production on triple sugar iron agar, fermentation of cellobiose, raffnose and adonitol negativity [7]. Out of 253 E. coli isolates 203 were identified as STEC by multiplex PCR as per the method of Paton and Paton 1998 [8] and 50 were non-STEC. Isolates possessing either stx1 or stx2 or both genes by multiplex PCR were considered as STEC and isolates without any genes were considered as non-STEC. Of 203 STEC isolates, 159 from raw meat, 19 from RTE meat foods, 18 from drinking water and 7 from human diarrhoeic samples. The STEC isolates from raw meat include 23 from buffalo meat, 21 from mutton, 38 from chevon, 41 from pork, and 21 from fish and 15 from chicken. The STEC isolates from RTE meat foods include, 7 from chicken products, 6 from mutton products, 4 from pork products and 1 from each chevon and buffalo meat products.
Antimicrobial susceptibility testing
A total of 253 E. coli (203 STEC and 50 STEC) isolates were tested for antibiotic resistance by standard disk diffusion technique [9] on Mueller Hinton agar using commercial discs (HiMedia, Mumbai, India). The following antibiotics with the disc strength in parentheses were used: amikacin (30 μg), amoxycillin (30 μg), ampicillin (10 μg), cefaclor (30 μg), cephotaxime (30 μg), chloramphenicol (30 μg), ciprofloxacin (5 μg), colistin (10 μg), co-trimoxazole (1.25/23.75 μg), erythromycin (15 μg), gentamycin (10 μg), kanamycin (30 μg), linezolid ( 30 μg), norfloxacin (10 μg), ofloxacin ( 5 μg), penicillin G (10 μg), polymyxin B (300 units), streptomycin (10 μg, tetracycline (30 μg) and trimethoprim (5 μg).
Cluster analysis
To evaluate the strain diversity of STEC isolates from different raw meat, RTE meat foods, water and human diarrhoeic samples, dendograms of antibiotic resistance patterns were constructed by using Treecon software.
Polymerase chain reaction for the detection of tetracycline and streptomycin resistance genes
Isolated bacterial colonies were suspended in 100 μl of sterilized distilled water. The suspensions were boiled, cooled and centrifuged, and the supernatants were used as template. All the PCR experiments were performed in 25μl volumes containing 3μl of prepared sample supernatant as template in a PCR mix consisting of 10mM Tris pH 8.3, 1.5 mM MgCl2, 50 mM KCl, 0.005% Tween 20, 0.005% Nonidet NP40 (Fermentas), 0.25μM of each primer and 200 μM of each dNTP, I U of Taq DNA polymerase. All amplifications consisted of an initial denaturation at 95°C for 4 min, followed by 35 cycles of denaturation at 95°C for 1 min, annealing at 58°C for tetA, tetB and tetC and 55°C for strA and strB for 1 min and elongation at 72°C for 1 min and final elongation at 72°C for 7 min. The primers were used for the detection of tetA, tetB, tetC, strA and strB genes were as per the method of Boerlin et al 2005 [10]. In each PCR reaction the positive and negative controls were included.
Cloning and sequencing of tetA and tetB genes of shiga toxin-producing E.coli O69 serotype of buffalo meat isolate
The E. coli O69 serotype was chosen for sequencing studies for the following reasons. Firstly, O69 is the most common STEC serotype recovered from buffalo meat and buffalo meat RTE products. Secondly the majority of the O69 serotypes carried both tetA and tetB tetracycline resistant determinants. The PCR amplicons (502 bp for tetA and 930 bp for tetB) were recovered from low melting point agarose, purified, and cloned with a pUCm-T vector. White colonies of ampicillin–resistant transformants were screened for the presence of tetA and tetB fragments by PCR with the same primer sets (TetA-F 5’GGCGGTCTTCTTCATCATGC3’,TetAR5’ CGGCAGGCAGAGCAAGTAGA3’and TetB-F5’CATTAATAGGCGCATCGCTG3’, TetB-R 5’ TGAAGGTCATCGATAGCAGG3’ ) used for amplification. DNA sequence of tetA and tetB genes were performed using the Big Dye terminator cycle sequencing method with Ampli Taq DNA polymerase in ABI PRIZM 377 DNA sequencer(Perkin-Elmer).The fragments were sequenced at least twice with each primer to reduce possibility of sequencing artefacts. Compilation and analysis of DNA sequence data were performed using Auto Assembler software 9Perkin- Elmer). Nucleotide and aminoacid homology analysis was performed using the BLAST (Basic Local Alignment Search Tool) in GenBank [11].
Nulceotide sequence accession number: The sequences of tetA and of O69 serotype of shiga toxin-producing E. coli of buffalo meat were submitted to the GenBank database and have been given the asscession numbers AF497970 and FJ7917423 respectively.
Results
Antibiotic resistance patterns
Of the 253 E. coli isolates used in this study, 203 (81.2%) were STEC and 50 (19.8%) were non STEC. To detect the resistance/ sensitivity pattern of STEC and non-STEC isolates from different sources, in vitro antibiotic sensitivity test was carried out by disk diffusion method using 20 commercially available Himedia (India) antibiotic discs. All the isolates of STEC and non- STEC were resistant to at least one of the antibiotics tested.
Globally, the level of resistance to each of the 20 antimicrobial agents assayed among STEC and non-STEC proved to be very similar.
Antimicrobial resistance patterns study revealed that STEC and non-STEC isolates were completely resistance to penicillin (253/253, 100%) followed by linezolid (249/253, 98.4%), erythromycin (245/253, 96.8%), streptomycin (232/253, 91.7%), tetracycline (223/253, 88.1%), ampicillin 123/253, 48%), cephotaxime (106/253, 41.9%), trimethoprim (93/253, 36.8%)%), cotrimoxazole (85/253, 33.6%), cefaclor (84/253, 33.2%), amoxycillin (79/253, 31.3%), ciprofloxacin (78/253, 30.8%), kanamycin (76/253, 30%), norfloxacin (62/253, 24.5%), ofloxacin (60/253, 23.7%), chloramphenicol (35/253, 13.8%), colistin (25/253,9.9%), polymyxin-B (27/253, 10.7%), amikacin (22/253, 8.7%) and gentamycin (17/253, 6.7%)
Out of 203 STEC isolates 73 (35.9%) isolates showed resistance to more than 50% of the antibiotics tested. Only one STEC isolate from pork showed resistance to 90% of the antibiotics tested.
On source wise analysis of the resistant STEC isolates 17 pork isolates, 16 goat meat isolates, 9 chicken isolates, 9 water isolates, 6 isolates from RTE meat foods, 6 human isolates, 5 fish isolates, 3 mutton isolates, and 2 isolates from buffalo meat showed resistance to 10 to 17 antibiotics tested.
Out of 50 non–STEC isolates, 24 (48%) showed resistance to more than 50% of the antibiotics tested. Only one isolate from humans showed resistant to 18 (90%) antibiotics tested. Among the 24 non – STEC isolates, 7 human isolates, 6 pork isolates, 4 RTE products isolates, 2 chicken isolates, 2 fish isolates, 1 goat meat isolate, 1 mutton isolate and 1 water isolate showed resistance to more than 50% of the antibiotics tested.
Sourcewise details of antibiotic resistance patterns of STEC and non-STEC are presented in Table 1 and 2. Of the 253 isolates from various sources in this study, the highest frequencies of antimicrobial resistance phenotypes were observed for STEC and non-STEC isolated from humans. When compared with the food isolates human STEC isolates were resistant to cefaclor, erythromycin, linezolid, pencillin G, streptomycin and tetracycline (100% each), ampicillin, amoxycillin, cephotaxime and kanamycin (85.7%), ciprofloxacin, norfloxacin, ofloxacin, trimethoprim and co-trimoxazole (71.4%), gentamycin (57.1%), polymyxin B (28.6%) and amikacin (8.9%).
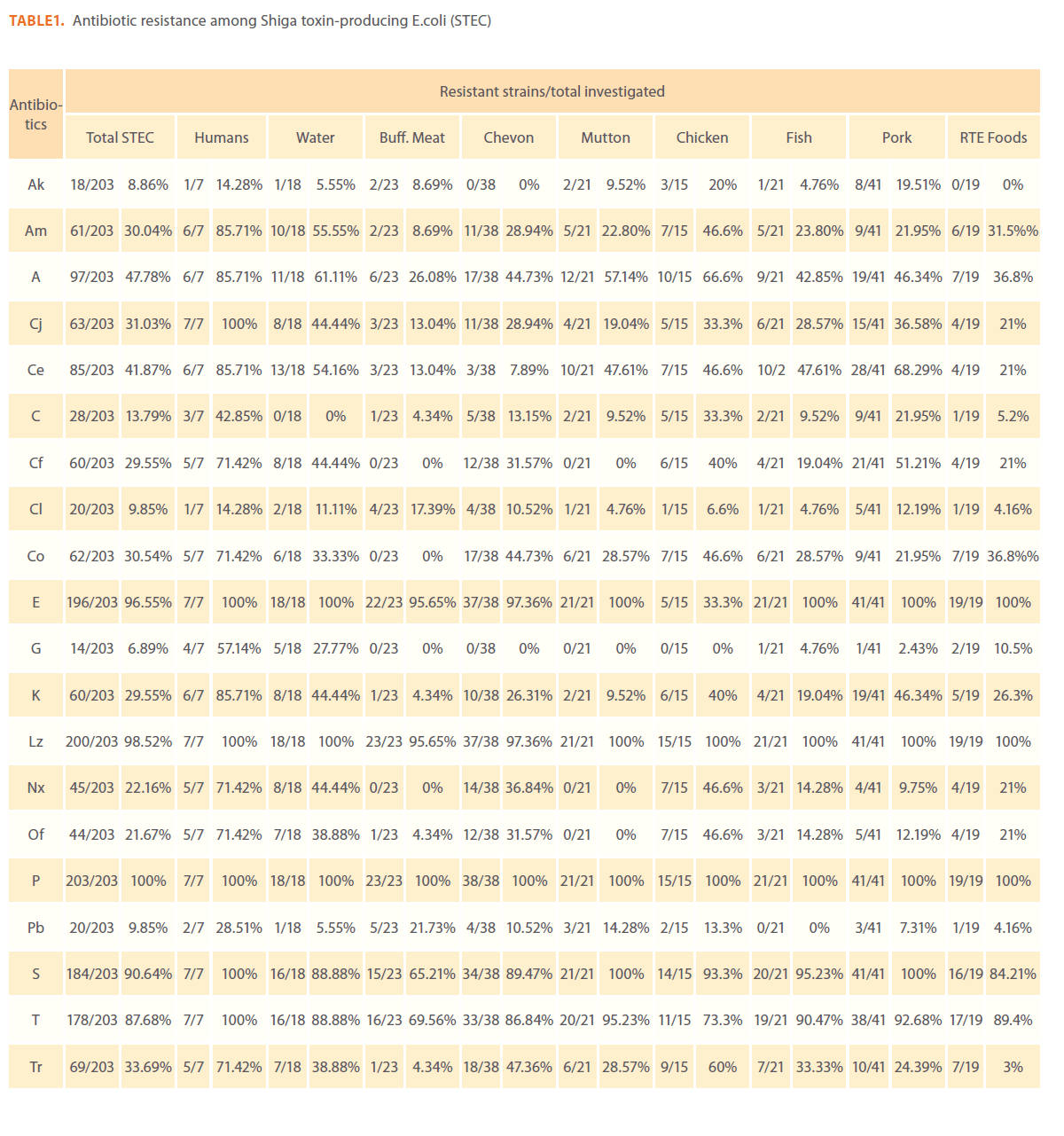
Table 1: Antibiotic resistance among Shiga toxin-producing E.coli (STEC)
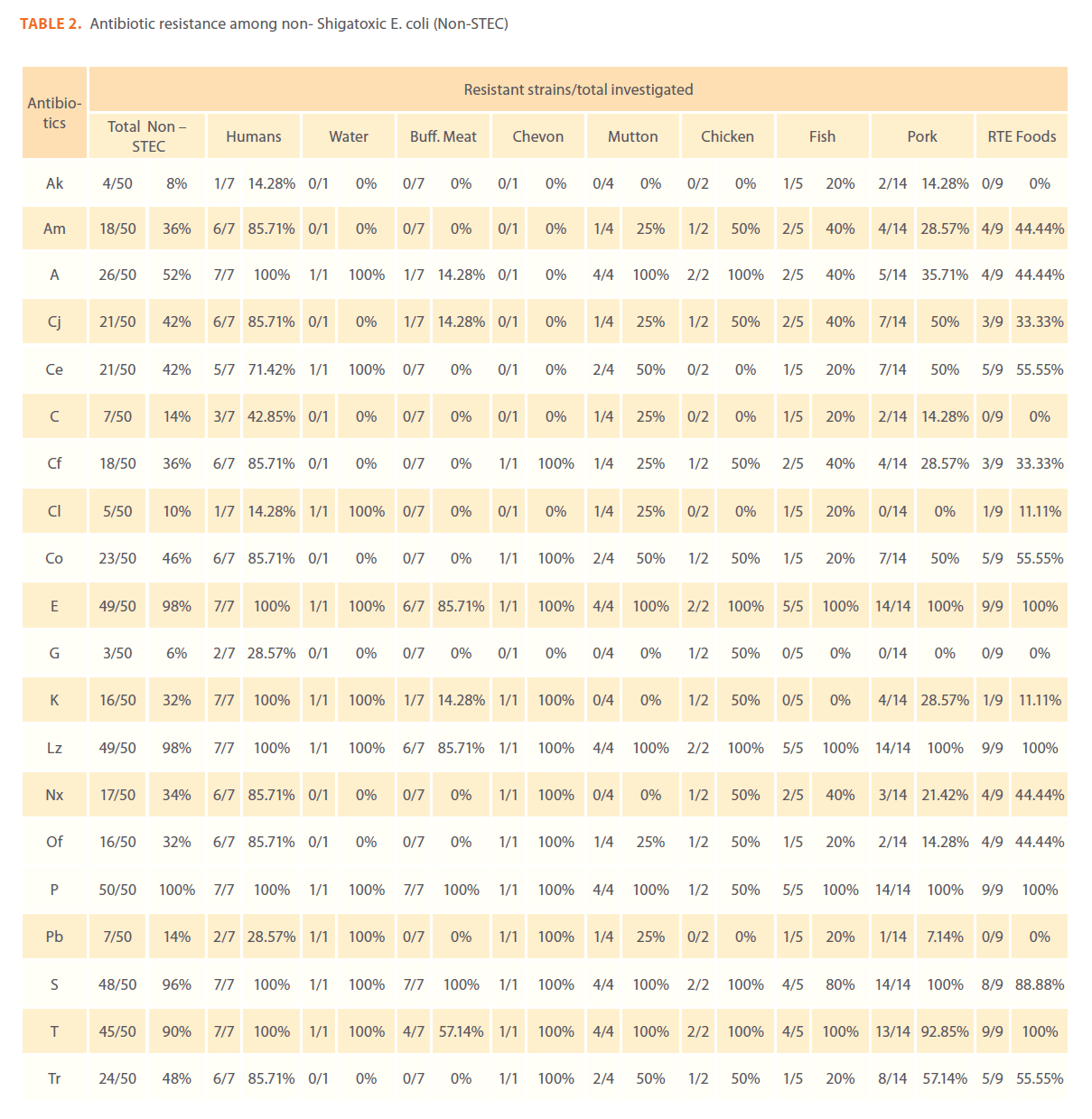
Table 2: Antibiotic resistance among non- Shigatoxic E. coli (Non-STEC)
Resistance profiles among STEC isolates from buffalo meat, chevon, pork and fish were almost similar to each other. Hundred percent of STEC isolates from buffalo meat were resistant to penicillin G followed by 95.7% to erythromycin and linezolid, 69.6% to tetracycline, 65.2% to streptomycin, 26.1% to ampicllin, 21.7% to polymyxin B and 17.4% to colistin. Hundred percent of STEC isolates from chicken were resistant to erythromycin, linezolid and penicillin G, 93.3% to streptomycin, 73.3% to tetracycline, 66.6% to ampicillin, 60% to trimethprim, and cephotaxime, 46.6% to amoxicillin, norfloxacin and ofloxacin and, 40% to ciprofloxacin and amikacin.
Cluster analysis
To evaluate the strain diversity of STEC isolates from different raw meat, RTE meat foods, water and human diarrhoeic samples, dendograms of antibiotic resistance patterns were constructed by using unweighted pair group method with arithmetic means (UPGMA) tree building and significant clusters in each dendogram were identified (Fig. 1 and 2) The dendogram revealed that STEC isolates from different sources of the study formed 4 significant clusters (Cluster I, cluster II, cluster III and minor formations).
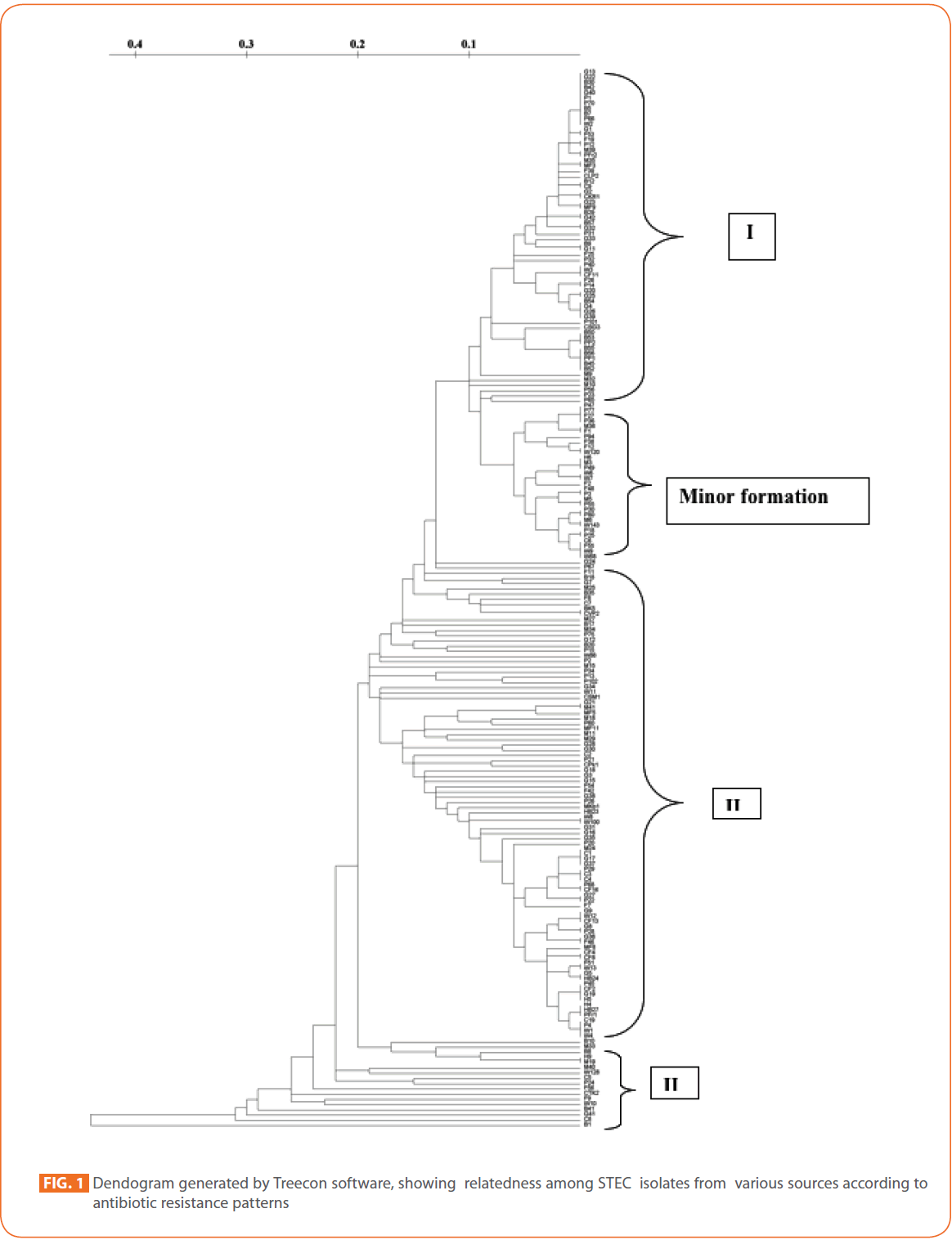
Figure 1: Dendogram generated by Treecon software, showing relatedness among STEC isolates from various sources according to antibiotic resistance patterns/p>
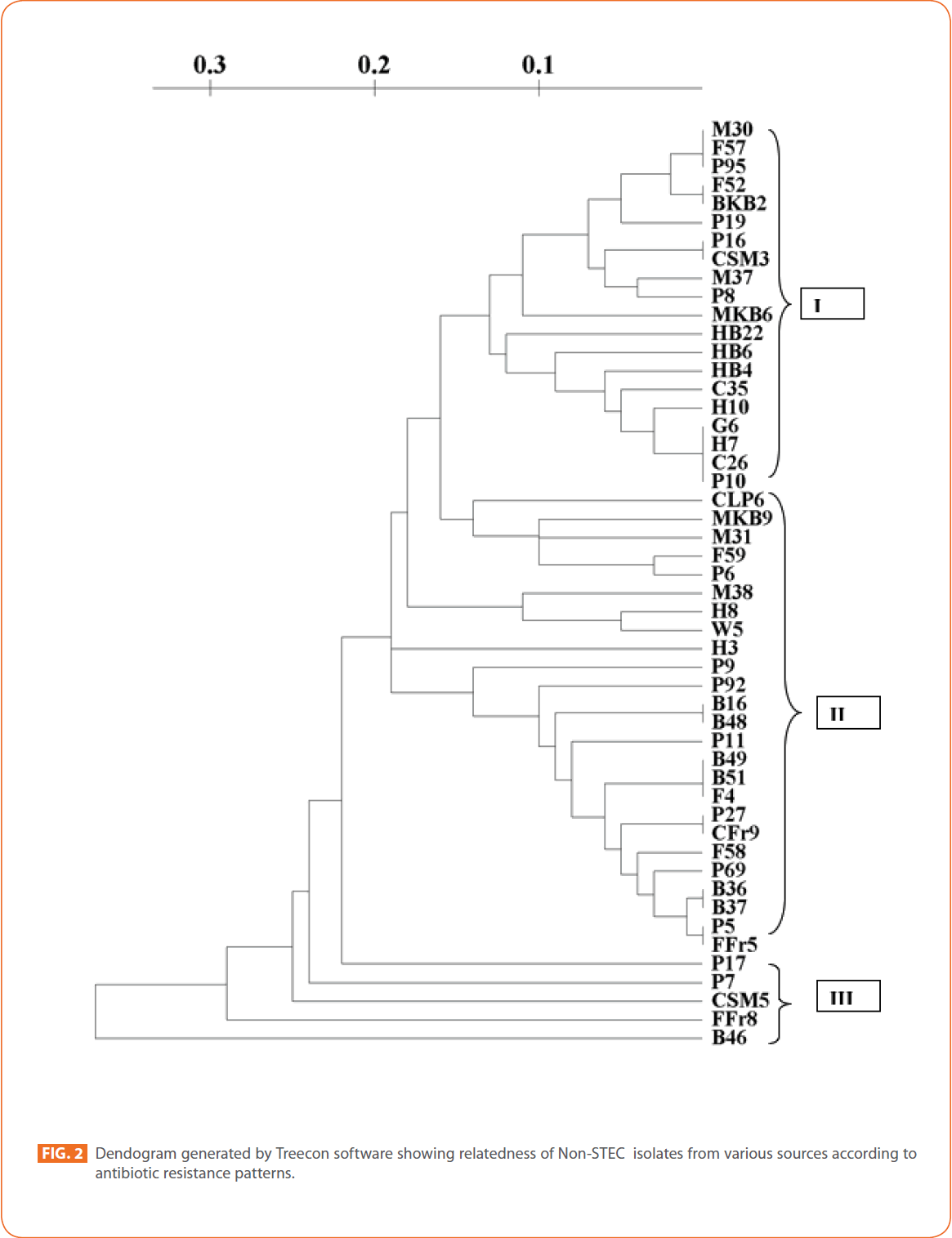
Figure 2: Dendogram generated by Treecon software showing relatedness of Non-STEC isolates from various sources according to antibiotic resistance patterns.
Cluster I comprised mainly STEC isolates of goat meat, buffalo meat and pork. Human STEC isolates were absent in cluster I.
In tightly knit cluster II, majority of goat meat isolates, pork isolates and RTE meat foods isolates were present. Some chicken, fish, mutton, humans and water isolates were also present.
Majority of the Pork isolates (15) formed as a separate group (minor formation). In cluster III, 17 isolates of different sources (Buffalo meat-4, mutton-3 water-2, fish-2, chicken-2, RTE meat foods-1, humans-1, chevon-1 and pork-1) were present.
Cluster/proximity analysis of non-STEC isolates from different sources revealed 3 significant clusters. Pork isolates and human isolates were the major non-STEC isolates in cluster I. Some isolates from fish, mutton, chicken, goat meat and RTE meat foods were also present. No isolates from water sources were present in cluster I.
In cluster II isolates from buffalo meat and pork were the major source. Isolates from water, fish, mutton RTE meat foods and humans were also present. Chicken isolates and goat meat isolates were absent in this cluster. In cluster III only pork and RTE meat foods isolates and one isolate from buffalo meat were present. Isolates from goat meat, water, fish, mutton, chicken and humans were absent in this cluster.
Distribution of tetA, tetB and tetC resistance genes
All 223 isolates (178 STEC and 45 non-STEC), which were phenotypically resistant to tetracycline by disk diffusion method were examined further by using PCR for the presence of tetA, tetB, tetC genes. The frequencies of the major resistance genes for tetracycline in the different E. coli isolates of different sources, buffalo meat, chevon, mutton, chicken, fish, humans, water and RTE meat foods are reported in Table. 3. Of the 178 shigatoxic E.coli showing phenotypic resistance to tetracycline analyzed by PCR, 177 (99.5%) STEC contained at least one of three (tetA, tetB, tetC ) tetracycline resistance genes (Fig. 3)
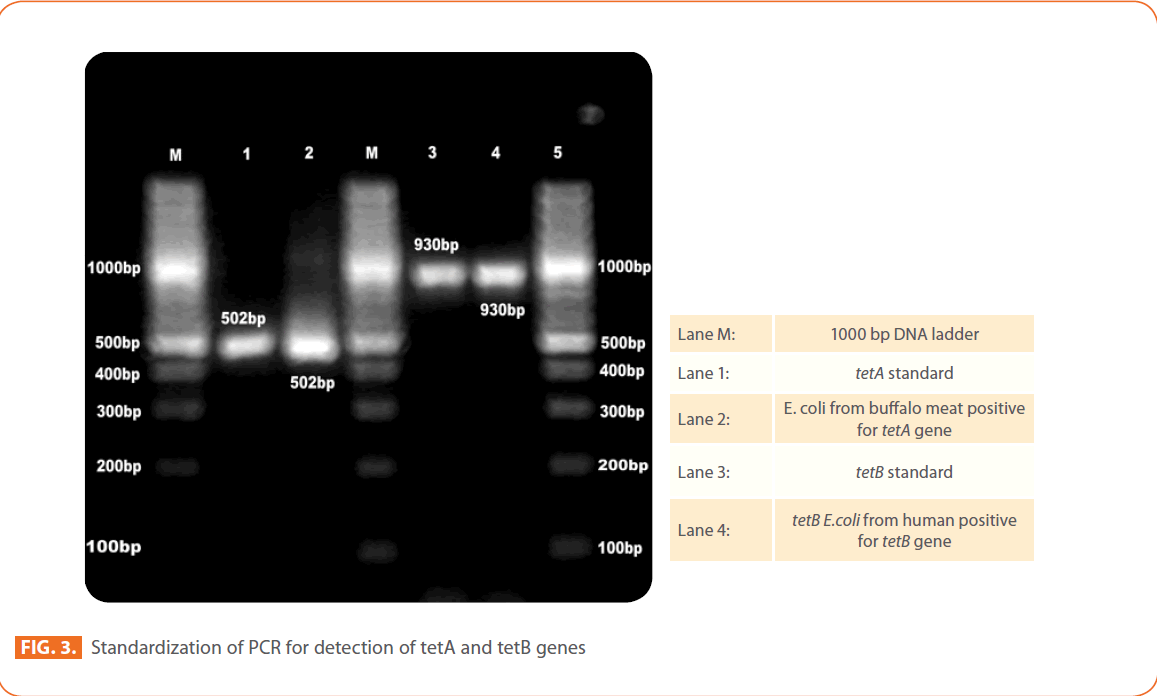
Figure 3: Standardization of PCR for detection of tetA and tetB genes
STEC isolates
The most common resistance genes were tetA (107 of 178; 60%) and tetB (48 of 178; 27%). Twelve per cent of STEC isolates were carried both tetA and tetB resistance genes. Only one isolate from chicken sausage has shown tetC resistance gene. Among STEC isolates tetA gene was predominant in humans (100%), chicken (86%), mutton (80%), Chevon (67%), Pork 63%), buffalo meat (62.5%) RTE meat foods (36%) and water (31%) STEC isolates.
The tetB resistance gene was present in 50% of water, 37% of fish, 32% of pork, 27% of chevon, 14% of chicken and 12.5% of buffalo meat STEC isolates. All human STEC isolates were negative for tetB determinants. Both tetA and tetB genes were present in 25% of buffalo meat, 19% of water, 18% of RTE meat foods STEC isolates. None of the human and chicken STEC isolates carried both tetA and tetB resistance genes in combination.
Non-STEC isolates
The tetA gene was predominant in all (100%) non-STEC isolates of buffalo meat, chevon, and water, 75% of mutton, 62% of pork, 56% of RTE meat foods, 43% of humans and 25% fish isolates. In chicken isolates tetA resistance gene was absent. The tetB resistance gene was present in 50% of chicken and fish, 23% of pork and 22% of RTE meat food isolates and was absent in buffalo meat, chevon, mutton, water and human isolates.
Fifty seven per cent of human, 50% of chicken, 25% of fish, 22% of RTE meat foods and 15% of pork non-STEC strains carried both tetA and tetB tetracycline resistance genes. Among 45 non-STEC isolates tested by PCR, 98% contained the tetracycline resistance genes. The most common resistance genes were tetA (58%), followed by tetA and tetB in combination (22%), and tetB (18%). All thenon STEC isolates were negative for tetC gene.
Out of 223 E. coli isolates phenotypically resistance to tetracycline, only one STEC isolate from fish and one non-STEC isolate from mutton lacked all three tetracycline resistance genes.
Distribution of strA and strB genes
All 231 isolates (184 STEC and 47 non-STEC), which were phenotypically resistant to streptomycin by disk diffusion method were examined further by using PCR for the presence of strA and strB genes (Fig. 4 and 5). The frequencies of the major resistance genes for streptomycin in the E. coli isolates of different sources, buffalo meat, chevon, mutton, chicken, fish, humans, water and RTE meat foods are reported in Table 3.
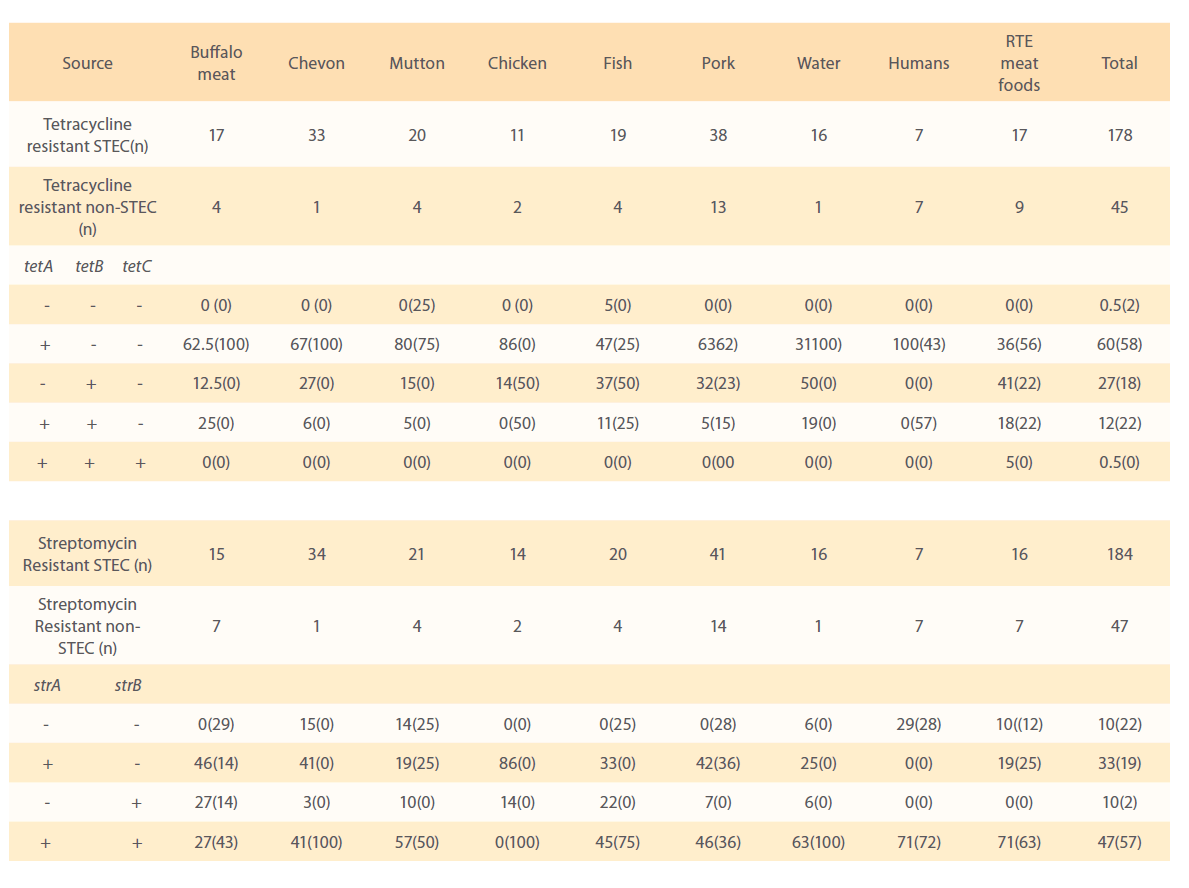
Figure 4: Standardization of PCR for detection of strA gene.
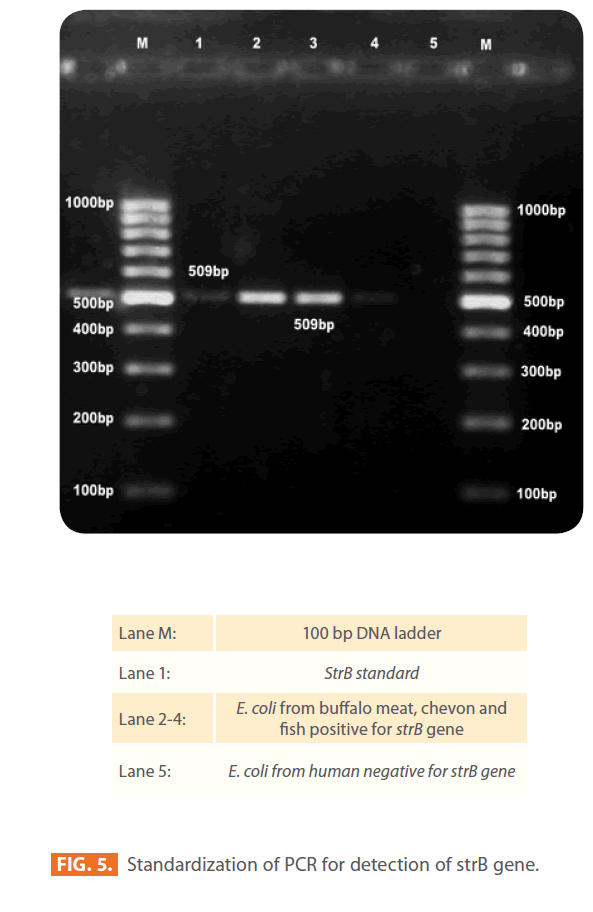
Figure 5: Standardization of PCR for detection of strB gene.
Of the 184 STEC isolates showing phenotypic resistance to streptomycin were investigated for strA and strB resistance genes, 86 (47%) of isolates harboured both strA and strB genes. Sixty (33%) and 18 (10%) isolates carried strA and strB gene, respectively and 18 (10%) of isolates did not carry any. Seventy one per cent of isolates from humans and RTE meat foods, 63% of water, 57% of mutton,46% of pork, 45% of fish, 41% of chevon and 27% from buffalo meat isolates harboured both strA and strB resistance genes. All 14 chicken isolates were negative for both strA and strB genes in combination. Eighty six percent of chicken, 46% of buffalo meat, 42% of pork, 41% of chevon, 33% fish, 25% of water, 19% of mutton and RTE meat food isolates carried only strA gene. Twenty seven per cent of buffalo meat, 22% of fish, 10% of mutton, 14% of chicken, 7% of pork, 6% water and 3% of chevon isolates carried strB resistance gene.
Of 47 non-STEC isolates analyzed for strA and strB genes, 57% isolates carried both strA and strB genes, 19% and 2% isolates carried strA and strB resistance gene respectively and 22% of the isolates did not carry any gene.
Nucleotide sequencing and accession numbers
The nucleotide sequencing of tetracycline resistance genes tetA and tetB of O69 serotype of STEC buffalo meat isolate determined in this study were submitted to the GenBank and have been given accession numbers AF497970 and FJ7917423. The nucleotide sequences of tetA and tetB of O69 serotype of STEC buffalo meat isolates were 99-100% similar to the other published sequences in GenBank.
Discussion
Of the 253 E.coli isolates (203 STEC and 50 non-STEC) in this study, approximately 90% displayed resistance to one or more antimicrobials including penicillin G, sulphonamides, tetracyclines, cephalosporins and aminoglycosides. The results in the present study are more or less similar to the earlier findings, where along with multiple resistance of E. coli to several antibiotics, a varying degree of sensitivity was reported for norfloxacin, ciprofloxacin, ofloxacin, gentamycin, chloramphenicol, amikacin, cefaclor, cephotaxime and polymyxin-B [10,12-14].
The results from this study show alarming resistant frequencies in E.coli (both STEC and non-STEC) from humans, drinking water, raw meats and RTE meat foods. Antibiotic resistance patterns in this study also revealed that clear variations among resistance patterns between human isolates and foods. These differences could be related to the different antibiotic regimes used for the different antimicrobial agents in livestock species and humans [15]. In the present investigation high antimicrobial resistance pattern is possibly as a consequence of extensive usage of these antibiotics in the treatment of livestock or in other sources e.g.;- contact with animal faeces, feeding, water sources, agriculture which may be a cause of the transmission of resistant genes from various sources to food producing animals. Some strains are resistant to more than 10 antimicrobial agents tested, is an alert for the consumers who eat improperly cooked meat and unhygienic handle of meat before and after preparation. Epidemiological studies are required to find out the incidence of STEC in different raw as well as RTE meat foods and major possible contamination sources including abattoir hygiene, transportation of carcass, butcher’s hygiene, utensils used in butchers shop and storage conditions of meat and meat products should be checked regularly. No study so far has been conducted to understand the effects of antibiotic use in animals and agriculture in India. The unregulated use of antibiotics in such systems in India contributes significantly to the antibiotic residues in meat and meat products. The antibiotic resistance patterns of different meats and meat products strains of both STEC and non-STEC observed in this study suggests a greater risk in the form of transfer of resistance to other pathogenic bacteria. The antibiotic resistance in non-STEC cannot be ruled out as insignificant since the transfer of resistance genes can takes place between closely related bacteria such as members of Enterobacteriaceae family.
The correlation between genotype (absence or presence of a resistance gene) and phenotype (sensitive or resistant) was high for tetracycline (98% agreement). In contrast to studies from other countries, where tetB and tetC have shown to be dominant [16-19], tetA was the frequent tetracycline determinant in the present study, where was tetC was very infrequent. However, this observation is consistent with the studies of Lanz et al [14] and Boerlin et al [10], who reported high frequency of tetA than tetB and tetC in E.coli isolated from pigs with oedema and diarrhoea. The results of present study also showed that 22% of non-STEC isolates and 12% of STEC isolates contained both tetA and tetB genes. This is in contrast to results from previous findings, in which only 3.5% [20] and 5.4% [21] of isolates of pigs, cattle and chicken had two genes. However results of the present study are in agreement with the findings of Bryan et al 2004 [16] who reported 22.2% E. coli isolates of pigs, chickens, turkeys, sheep, cow, fish, goat, humans, dogs, cats, horses and goose, duck and deer contained two genes. Out of 223 isolates phenotypically resistance to tetracycline, only one STEC isolate from fish and one non-STEC isolate from mutton lacked all three tetracycline resistance genes. The reason for absence of all three resistance genes may be that in isolates point mutations may be causing phenotypic tetracycline resistance [22] or phenotypic resistance may be due to other plasmid mediated resistance determinants like tetG or chromosomal mediated tetM genes, which are not tested in the present study.
In this study 47% of STEC isolates and 57% of non-STEC isolates contained both strA and strB resistance genes; this finding is in agreement with the results of other researchers showing that both genes have to be present in order to obtain a functional streptomycin resistance [23]. Nineteen per cent of STEC and 33% of non-STEC isolates carried strA gene and 10% of STEC and 2% of non-STEC isolates carried strB gene only, in these isolates an additional aadA gene (The aadA gene did not tested in the present study) could be responsible for observed resistance phenotype. Ten per cent of STEC and 22% of non-STEC were lacked strA or strB or in combination, one step resistance mutations is the most probable explanation for resistance in these isolates [22]. Due to paucity of similar data, cluster analysis results could not be compared.
In conclusion, our work identified and compared for the first time resistance phenotypes and genotypes of STEC and non- STEC isolated from different raw meat, RTE meat foods, drinking water and human sources. We could show, that significance differences with regard to mutidrug resistance patterns can be observed between STEC and non-STEC isolates of different sources; raw meat, RTE meat products, drinking water and human diarrhoeic samples not only at the phenotypic level but also at the genotypic level for tetracycline and streptomycin. A comprehensive surveillance is required to determine the presence and distribution of antibiotic resistant E. coli in foods of animal origin and agriculture in India to monitor any emerging antimicrobial resistance problems that may arise in this important human pathogen.
Acknowledgements
We are grateful to Dr. Patrick Boerlin Department of Pathobiology, University of Guelph, Canada for kindly gifted DNA standards for the detection of tetA, tetB and tetC genes. We also thankful to the GADVASU authorities for providing necessary facilities and financial help for this study.
219
References
- Sorum H, Sunde M (2001) Resistance to antibiotics in the normal flora of animals. Vet Res 32: 227-241.
- Skurnik D et al. (2006) Effect of human vicinity on antimicrobial resistance and integrons in animal faecal Escherichia coli. J Antimicrob Chemother 57: 1215-1219.
- Johnson JR, Kuskowski MA, Smith K, O’Bryan TT, Tatini S (2005) Antimicrobial-resistant and extraintestinal pathogenic Escherichia coli in retail foods. J Infect Dis 191: 1040-1049.
- Neu HC (1992) The crisis in antibiotic resistance. Science 257: 1064-1073.
- Witte W (1998) Medical consequences of antibiotic use in agriculture. Science 257: 1064-1073.
- Braoudaki MaH, A, C (2003) 5th Int. Symp. Shiga toxin (verocytotoxin)-producing Escherichia coli infections. In: Edinburgh, UK,
- Edwards P, R. and Ewing , W, H (1972) Identification of enterobactriaceae. 4th eds. Elsvier Science, Burgees Publishing Co., USA.:
- Paton AW, Paton JC (2002) Direct detection and characterization of Shiga toxigenic Escherichia coli by multiplex PCR for stx1, stx2, eae, ehxA, and saa. J Clin Microbiol 40: 271-274.
- Bauer AW, Kirby WM, Sherris JC, Turck M (1966) Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol 45: 493-496.
- Boerlin P et al. (2005) Antimicrobial resistance and virulence genes of Escherichia coli isolates from swine in Ontario. Appl Environ Microbiol 71: 6753-6761.
- Madden TL, Tatusov RL, Zhang J (1996) Applications of network BLAST server. Methods Enzymol 266: 131-141.
- Guerra B, Junker E, Schroeter A, Malorny B, Lehmann S, Helmuth R (2003) Phenotypic and genotypic characterization of antimicrobial resistance in German Escherichia coli isolates from cattle, swine and poultry. J Antimicrob Chemother 52: 489-492.
- Karama M, Johnson RP, Holtslander R, McEwen SA, Gyles CL (2008) Prevalence and characterization of verotoxin-producing Escherichia coli (VTEC) in cattle from an Ontario abattoir. Can J Vet Res 72: 297-302.
- Lanz R, Kuhnert P, Boerlin P (2003) Antimicrobial resistance and resistance gene determinants in clinical Escherichia coli from different animal species in Switzerland. Vet Microbiol 91: 73-84.
- Schwarz S C-DE (2001) Use of antimicrobials in veterinary medicine and mechanisms of resistance. Vet Res 32: 201-225.
- Bryan A, Shapir N, Sadowsky MJ (2004) Frequency and distribution of tetracycline resistance genes in genetically diverse, nonselected, and nonclinical Escherichia coli strains isolated from diverse human and animal sources. Appl Environ Microbiol 70: 2503-2507.
- Roberts MC (2002) Resistance to tetracycline, macrolide-lincosamide-streptogramin, trimethoprim, and sulfonamide drug classes. Mol Biotechnol 20: 261-283.
- Srinivasan V et al. (2007) Phenotypic and genotypic antimicrobial resistance patterns of Escherichia coli isolated from dairy cows with mastitis. Vet Microbiol 124: 319-328.
- Sunde M (2005) Prevalence and characterization of class 1 and class 2 integrons in Escherichia coli isolated from meat and meat products of Norwegian origin. J Antimicrob Chemother 56: 1019- 1024.
- Marshall B TC, Levy SB. (1983) Frequency of tetracycline resistance determinant classes among lactose-fermenting coliforms. Antimicrob Agents Chemother. 24(6): 835-840.
- Sengelov G, Halling-Sorensen B, Aarestrup FM (2003) Susceptibility of Escherichia coli and Enterococcus faecium isolated from pigs and broiler chickens to tetracycline degradation products and distribution of tetracycline resistance determinants in E. coli from food animals. Vet Microbiol 95: 91- 101.
- Chopra I, Roberts M (2001) Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol Rev 65: 232-260 ; second page, table of contents.
- Chiou CS, Jones AL (1995) Expression and identification of the strA-strB gene pair from streptomycin-resistant Erwinia amylovora. Gene 152: 47-51.













