Ibanga IA1*, Akan OD1,2, Uyanga FZ1,3, Mantu EC4 and Asuquo M1
1Microbiology Department, Akwa-Ibom State University, Mkpat Enin LGA, Akwa-Ibom State, Nigeria
2College of Food Science and Technology, Central South University of Forestry and Technology, Hunan, PRC
3Department of Microbiology, Michael Okpara University of Agriculture, Umudike, Abia State, Nigeria
4Department of Microbiology, Federal School of Medical Laboratory Technology (Sciences), Jos, Plateau State, Nigeria
- *Corresponding Author:
- Ibanga IA
Microbiology Department, Akwa-Ibom State University
Mkpat Enin LGA, Akwa-Ibom State, Nigeria
E-mail: inimfonibanga@aksu.edu.ng
Received Date: August 08, 2019; Accepted Date: February 21, 2020; Published Date: February 27, 2020
Citation: Ibanga IA, Akan OD, Uyanga FZ, Mantu EC, Asuquo M (2020) Multi-Drug Resistance Pattern of Staphylococcus aureus from Paediatric ward, General Hospital, Ikot-Ekpaw, Mkpat Enin LGA, Akwa-Ibom State, Nigeria. Arch Clin Microbiol Vol. 11 No. 1:105.
DOI: 10.36648/1989-8436.11.1.105
Keywords
Multi-drug resistance; Staphylococcus aureus; Multiple antibiotics resistance index; Susceptibility test; Nosocomial infections
Introduction
Microbes do manifest themselves in three ways, through substance spoilage, fermentation of organic and inorganic matters and causation of ailments. Different microorganisms, with their different mode of aetiology, causing different types of ailments, will require different methods and capable drugs for treatments. Continuous deployment of antimicrobial drugs in treating microbial infections has led to the emergence of resistance amongst various strains of microorganisms [1,2]. MDR literally means ‘ being resistant to more than one antimicrobial agent’, although a standardized definition has not yet been agreed upon by the medical community. There are currently other definitions that are used to characterize patterns multidrug resistance.
The most practical definition used for Gram-positive and Gram-negative bacteria is ‘ resistance to three or more antimicrobial classes according to Magiorakos et al. [3]. MDR could also be defined as the insensitivity or resistance of a microorganism to administered antimicrobial medicines (which are structurally unrelated and have different molecular targets) despite earlier sensitivity to the same medicines [4]. According to Nikaido [5], multidrug resistance in bacteria cells come about by their accumulation, on resistance (R) plasmids or transposons, of genes, with each coding for resistance to a specific agent, and/or by the action of multidrug efflux pumps, each of which can pump out more than one drug type. This MDR abilities lead to ineffective ailment treatment, resulting in its persistence, infection’s spread and high cost [2,6].
The hospital environment have been said to be an active reservoir for infectious microorganisms, being the meeting point for people with diverse disease etiological agents and susceptible individuals [7-9]. Nikaido [5] mentioned that Staphylococcus aureus has a known nosocomial, multi-drugresistant strain referred to as the methicillin-resistant Staphylococcus aureus (MRSA).
MRSA was initially controlled but currently is also resistant to other antimicrobials like the aminoglycosides, chloramphenicol, lincosamides, macrolides and tetracycline [1]. This study was conducted as part of evidence to buttress the efficacy of Staphylococcal infections in young children and the scale to which MDR pathogens are becoming threats to the health of the younger generation amongst the Mkpat Enin, Akwa-Ibom State populace.
Materials and Methods
Study facility, group and sample collection
The study facility is in a growing town and services a couple of adjoining communities, with a number of established institutions, stable commerce and ever growing population. The General Hospital, Ikot-Ekpaw, in Mkpat Enin LGA, Akwa- Ibom State, South-South Nigeria, has Out-patient department, Post-natal ward, Pediatric ward and servicing laboratories. The study focused on children aged 1-15 years.
A hundred (100) sterile swabs samples from skin, nostrils, wound, throat and ear was sourced over a period of three months. Once gotten, the swabs were labelled, placed in an ice pack and taken to the Microbiology laboratory, Akwa-Ibom State University.
Sample analysis
The sterile swab sticks were depth into peptone water and incubated at 37oC for 24 hours. A loop full from each sample was streaked on separate Mannitol salt agar (MSA) plates and incubated at 37°C for 24 hours. Discrete golden yellow colonies were sub-cultured, purified and preserved. Only Gram positive cocci bacterial colonies were further tested for catalase and coagulase.
Antibiotics susceptibility test
Confirmed Staphylococcus aureus isolates were tested for their susceptibilities to various selected commercial antibiotic drugs like Ciprofloxacin, Erythromycin, Levofloxacin, Gentamicin, Ampiclox, Rifampicin, Amoxicillin, Streptomycin, Norfloxacin and Chloramphenicol. Overnight cultures using Kirby-Bauer method [10] were inoculated on Mueller-Hinton agar (Oxoid, Uk), cultures adjusted to 0.5 McFarland standard. After pre-diffusion, the plates were inoculated at 37°C for 24 hours.
Diameter of zones of inhibition (IZDs) produced by the antibiotics were measured and recorded in millimeter. Thereafter, the Multiple antibiotics resistance (MAR) index was determine for each isolate using a formula MAR = x/y, where x is the number of antibiotics to which test isolate displayed resistance and y is the total number of antibiotics to which the test organism has been evaluated for sensitivity [11,12].
Results
Result for the prevalence of S. aureus is as shown in Table 1. The result table shows that of the 28 confirmed, wound samples had the highest number, which was followed by samples from children skin swabs. The least number of confirmed Staphylococcus aureus isolates were from the ear swabs.
Table 1 Prevalence of Staphylococcus aureus from clinical samples.
| Type of Specimen |
Sample Size |
Number of S. aureus isolated |
Total percentage |
| Skin |
20 |
8 |
29 |
| Nose |
20 |
5 |
18 |
| Wound |
20 |
9 |
32 |
| Throat |
20 |
4 |
14 |
| Ear |
20 |
2 |
7 |
The susceptibility pattern of the 28 confirmed test isolates to the selected commercially available drugs (amoxicillin, ampiclox, chloramphenicol, ciprofloxacin, erythromycin, gentamicin, levofloxacin, norfloxacin, rifampicin and streptomycin) is as shown in Figure 1.
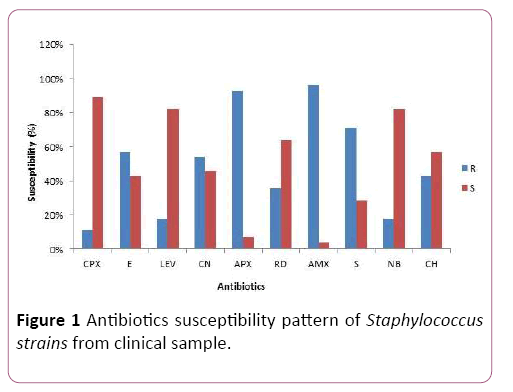
Figure 1: Antibiotics susceptibility pattern of Staphylococcus strains from clinical sample.
Ciprofloxacin was the most effective drugs against the test organism, followed by Levofloxacin and Norfloxacin. Confirmed Staphylococcus aureus isolates had high resistance for Amoxicillin, closely followed by their resistance for Ampiclox.
Table 2 has the multiple antibiotics resistance (MAR) index result which shows that 85.7% of the confirmed test isolates were multi-drug resistant (showing resistance to three or more classes of antibiotics). Only 14.3% of the isolates showed resistance to only two classes of antibiotics.
Table 2 Antibiotic Resistance Pattern and MAR index of Staphylococcus aureus.
| S/N |
Isolate code |
Antibiotic Resistant Pattern |
MARI |
Antibiotic Resistant Class |
| 1 |
S9 |
RD, S |
0.2 |
RIF, AMG |
| 2 |
S8 |
APX, AMX, CH |
0.3 |
PEN, CHL |
| 3 |
N3 |
LEV, APX, AMX |
0.3 |
QUI, PEN |
| 4 |
W1 |
CN, AMX, CH |
0.3 |
AMG, PEN, CHL |
| 5 |
W9 |
APX, AMX, S |
0.3 |
PEN, AMG |
| 6 |
S10 |
CN, APX, RD, AMX |
0.4 |
AMG, PEN, RIF |
| 7 |
T6 |
E, APX,AMX, NB |
0.4 |
MAC, PEN, QUI |
| 8 |
T8 |
CPX, E, APX, AMX |
0.4 |
QUI, MAC, PEN |
| 9 |
S2 |
E, APX, RD, AMX, S |
0.5 |
MAC, PEN, RIF, AMG |
| 10 |
S3 |
E, CN, APX, AMX, S |
0.5 |
MAC, AMG, PEN |
| 11 |
N1 |
E, CN, APX, AMX, CH |
0.5 |
MAC, AMG, PEN, CHL |
| 12 |
N2 |
CN, APX, AMX, S, CH |
0.5 |
AMG, PEN, CHL |
| 13 |
W2 |
E, LEV, APX, AMX, S |
0.5 |
MAC, QUI, PEN, AMG |
| 14 |
W5 |
E, APX, AMX, S, CH |
0.5 |
MAC, PEN, AMG, CHL |
| 15 |
W6 |
CN, APX, AMX, S, CH |
0.5 |
AMG, PEN, CHL |
| 16 |
W7 |
APX, AMX, S, NB, CH |
0.5 |
PEN, AMG, QUI, CHL |
| 17 |
W10 |
E, LEV, APX, AMX, S |
0.5 |
MAC, QUI, PEN, AMG |
| 18 |
T3 |
E, APX, AMX, S, CH |
0.5 |
MAC, PEN, AMG, CHL |
| 19 |
E5 |
E, CN, APX, RD, AMX |
0.5 |
MAC, AMG, PEN, RIF |
| 20 |
S1 |
E, CN, APX, RD, AMX, S |
0.6 |
MAC, AMG, PEN, RIF |
| 21 |
S5 |
E, LEV, APX, RD, AMX, S |
0.6 |
MAC, QUI, PEN, RIF,AMG |
| 22 |
S6 |
CN, APX, RD, AMX, S, CH |
0.6 |
AMG, PEN, RIF, CHL |
| 23 |
N5 |
CPX, CN, APX, AMX, S, CH |
0.6 |
QUI, AMG, PEN, CHL |
| 24 |
W8 |
E, CN, APX, AMX, S, NB |
0.6 |
MAC, AMG, PEN, QUI |
| 25 |
N8 |
E, CN, APX, RD, AMX, S, NB |
0.7 |
MAC, AMG, PEN, RIF, QUI |
| 26 |
W3 |
LEV, CN, APX, RD, AMX, S, NB |
0.7 |
QUI, AMG, PEN, RIF |
| 27 |
T2 |
E, CN, APX, RD, AMX, S, CH |
0.7 |
MAC, AMG, PEN, RIF, CHL |
| 28 |
E9 |
CPX, E, CN, APX, AMX, S, CH |
0.7 |
QUI, MAC, AMG, PEN,CHL |
KEY: CPX: Ciprofloxacin; E: Erythromycin; LEV: Levofloxacin; CN: Gentamicin; APX: Ampiclox; RD: Rifampicin; AMX: Amoxicillin; S: Streptomycin; NB: Norfloxacin; CH: Chloramphenicol; MARI: Multiple Antibiotic Resistance Index; RIF: Rifamycins; AMG: Aminoglycosides; PEN: Penicillins; CHL: Chloramphenicol; MAC: Macrolides; QUI: Quinolones
Results from this study show that 60.7% of test isolates had resistance for four or more antibiotics drugs. MAR index value for 96.4% of the test isolates reveal cases of source drug abuse.
Discussions and Conclusion
Data for isolate occurrence and confirmation showed that there were more confirmed staphylococcus isolates from wound swab-samples, followed by skin sample-swabs. Parta et al. [13] also recorded very high Staphylococcus number from wound swabs. This high isolate-numerical is suggestive of the exposed nature of the sampling points. This is supported by findings presented by Nimmo et al. [14], who found more Staphylococcus isolates on exposed body surfaces than the internal parts. While uncovered wounds have sticky surfaces and the skin is continuously exposed, it is therefore easy for such high microbial numbers to be recorded.
Susceptibility data showed that all the confirmed and tested isolates resisted two or more antimicrobials. Qureshi et al. [15] also isolated MRSA that resisted multiple anti-microbials from hospital specimens. This study’s result showed a higher MDR percentage than the “ nearly half ” proportion reported by Nimmo et al. [14].
More than 96.4% of the MAR indices were 0.3 from this study evaluation. This assertion is indicative that resistance to these multiple drugs come from over exposure of the isolates to drugs, making them adapt or resistant to them with recurrent treatments. The high case of MDR amongst microorganisms can drastically be reduced by mere restricting the indiscriminate and readily availability of these drugs over the counter [14].
Many studies have shown that prolonged stays in hospitals increases the risk for colonization or infection with MDR Staphylococcus aureus. This reflects an inherent risk in acquiring MDR organisms through environmental contaminations and hospital stay conditions. With infant patients, another potential transmission route is through infected staff members handling and this calls for special care [16,17].
The isolation of MDR Staphylococcus aureus from infant patients is a call for the proper implementation of contact precautions during hospitalization especially in developing countries [16,18].
Ethical Consideration
The study was approved by the ministry of health, Akwa Ibom State. Permission was obtained from General Administration of the General hospital prior to collecting any data. Participants ’ privacy and confidentiality have been assured (no names have been used, only numbers were used) and all data and results have been handled and treated confidentially.
26522
References
- McIntosh J (2018) Antibiotic resistance: What you need to know. Medical news today.
- Tanwar J, Das S, Zeeshan F, Hameed S (2014) Multidrug Resistance: An Emerging Crisis. Interdisciplinary Perspectives on Infectious Diseases.
- Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, et al. (2012) Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clinical Microbiology and Infection 18: 268-281.
- Singh V (2013) Antimicrobial resistanceIn:Microbial Pathogens and Strategies for Combating Them: Science, Technology and Education, FormatexResearchCenter 1: 291-296.
- Nikaido H (2009) Multidrug Resistance in Bacteria. Annual Review of Biochemistry 78: 119-146.
- WHO (2014) Antimicrobial Resistance Global Report on Surveillance. Geneva, Switzerland: World Health Organization.
- Zhannel GG, Corby D, Laing N, Weshionweski B, Vashisht R, et al.. (2008) Canadian Antimicrobial Resistant Pathogens in Intensive care Unit (CAN-ICU) study 2005-2006. Antimicrobial Agents and Chemotherapy 52: 1430-1437.
- Rhomberg PR, Fristoche TR, Sader HS, Jones RN (2006) Antimicrobial susceptibility pattern comparisons among intensive care unit and general ward Gram negative isolates from meropenem yearly susceptibility test Information programme (USA). Diagnostic Microbial Infectious Disease 56: 57-62.
- O’Brien FG, Pearman JW, Gracey M, Riley TV, Grubb WB (1999) Community strain of methicillin-resistant Staphylococcus aureus involved in a hospital outbreak. Journal of Clinical Microbiology 37: 2858-2862.
- Hudzicki J (2009) Kirby-Bauer disk diffusion susceptibility test protocol. American Society for Microbiology.
- Tula-Sanchez AA, Havas AP, Alonge PJ, Klein ME, Doctor SR, et al. (2013) A model of sensitivity and resistance to histone deacetylase inhibitors in diffuse large B cell lymphoma: Role of cyclin-dependent kinase inhibitors. Cancer Biology and Therapy 14: 949-961.
- Olayinka AT, Onile BA, Olayinka BO (2004) Prevalence of multi-drug resistance (MDR) Pseudomonas aeruginosa isolates in surgical units of Ahmadu Bello University Teaching Hospital, Zaria, Nigeria: An indication for effective control measures. Annals of African Medicine 3: 13-16.
- Parta M, Goebel M, Matloobi M, Stager C, Musher DM (2009) Identification of Methicillin-Resistant or Methicillin-SusceptibleStaphylococcus aureusin Blood Cultures andWound Swabs by GeneXpert. Journal of Clinical Microbiology 47: 1609-1610.
- Nimmo GR, Pearson JC, Collignon PJ, Christiansen KJ, Coombs GW, et al. (2011) Antimicrobial susceptibility of Staphylococcus aureus isolated from hospital in-patient. Community DiseaseIntelligence 35: 237-243.
- Qureshi AH, Rafi S, Qureshi SM, Ali AM (2004)The Current Susceptibility Patterns of Methicillin Resistant Staphylococcusaureus to Conventional Anti-Staphylococcus Antimicrobials at Rawalpindi. Pakistan Journal of Medical Science 20: 361-364.
- Buke C, Armand-Lefevre L, Lolom I, Guerinot W, Deblangy C, et al. (2007) Epidemiology of multidrug-resistant bacteria in patients with long hospital stays. Infection Control and Hospital Epidemiology 28: 1255-1260.
- Maamar E, Ferjani S, Jendoubi A, Hammami S, Hamzaoui Z, et al. (2016) High prevalence of gut microbiota colonization with broad-spectrum cephalosporin resistant enterobacteriaceae in a tunisian intensive care unit. Frontiers in Microbiology.
- Harbarth S, Sax H, Fankhauser-Rodriguez C, Schrenzel J, Agostinho A et al. (2006) Evaluating the probability of previously unknown carriage of MRSA at hospital admission. American Journal of Medicine 119: 275.e15-e23.






