Keywords
EDR peptide; Cell culture; Huntington’s disease; DNA-peptide interactions
Introduction
Huntington’s disease (HD) is a fatal, inherited neurodegenerative disorder. HD is caused by expansion of a cytosine-adenine-guanine (CAG) repeat in the coding region of the huntingtin gene (HTT), located on chromosome 4p16.3 [1]. The CAG repeats in HTT are translated into a polyglutamine (polyQ) sequence in the N-terminal region of the huntingtin (Htt) protein. HD typically occurs in midlife, but extensive CAG expansion leads to a juvenile onset of the disease. An average number of repeats makes 19 CAG in the huntingtin gene of unaffected individuals, while HD patients reveal 36 to 121 CAG repeats [2]. Patients with 36 to 39 repeats show reduced penetrance for the disease and can be asymptomatic for many years [3,4]. The most HD affected appears to be striatum with subsequent atrophy of the cerebral cortex at the late stage [5]. Mutant huntingtin (mHtt) is inclined to aggregation with its possible toxic effect on neurons. Mutant Htt induces neuronal cell death in several ways: gene transcription, formation of toxic aggregates, direct induction of apoptosis, disruption of key neuronal functions such as proteosomal or mitochondrial functions, ubiquitination pathways, axonal transport, endocytosis and synaptic transmission [6–9].
The mutant huntingtin protein was shown to directly and specifically bind to the inositol-1,4,5-triphosphate receptors type 1 (InsP3R1) and activate these receptors in lipid bilayers [10]. An increased release of Ca2+ from intracellular storage - endoplasmic reticulum - in striatal medium spiny neurons (MSN) in primary cultures of transgenic mice (models of HD), obviously causes cell death [11-14]. Expansion of polyQ sequence led to violation of Htt conformation thus resulting in Htt aggregation. It hindered the movement of vesicles containing neurotransmitters via the cytoskeleton, which disrupted signaling in neurons [15]. The search for effective treatment of HD is particularly relevant. Neurons functioning and aging studies may prompt new opportunities for regulation of these processes. Short neuroprotective peptides employment opens great perspectives for the development of novel highly effective and side effects free neuroprotective drugs aimed at treatment of neurodegenerative diseases.
Among short peptides showing potential modulatory functions is EDR (Glu-Asp-Arg) with pronounced neuro-protective properties [16].
EDR influence on the functional activity of the central nervous system was studied in an experimental model of prenatal hyper-homocysteinemia in rats. The induction of oxidative stress in vivo is known to be associated with a high level of homocysteine in the blood of animals, reduction of cognitive abilities and impairment of glutamatergic systems in the brain. Intramuscular injection of peptide EDR to rats contributed to the improvement of spatial orientation and learning capacity in progeny during “Morris water maze” test [16]. EDR protective effect might be related to its ability to inhibit the accumulation of reactive oxygen species (ROS) in the neurons, elevating their resistance to oxidative stress and preventing interaction between homocysteine and its derivatives with glutamate receptors [16]. Peptide EDR administration in the model of hypoxia in the embryonic or prenatal period of rats contributed to the restoration of the ability to latent learning up to a normal level in 3-week-old rats. The rats, prenatally administered with peptide EDR simulating a prenatal stress showed a normalized behavioral sleep, eating and relaxed waking [17].
The effect of peptide EDR on activation of MAP-kinase was evaluated in cultures of cerebellar granule cells. Its temporary profile determined which genes were going to be expressed - adaptation or apoptosis. Addition of peptide EDR in cell culture increased lag-time of MAP-kinase activation that could be estimated as a protective effect against homocysteine toxic action. The influence of peptide EDR on oxidation process caused by ouabain or hydrogen peroxide in neurons was recently investigated. Peptide EDR reduced ROS in neurons [18].
There is a range of peptides known to penetrate cells and concentrate on the surface of cell nucleus. As a rule, they have an enhanced content of arginine and lysine residues in their sequence [19,20]. Moreover, these peptides can not only penetrate into the cell, but also form complexes with DNA and RNA [21,22]. An important experimental fact that confirms the ability of short peptides to penetrate into the cell consists in registering FITC-labeled di-, tri-, and tetrapeptides penetration not only into the cytoplasm, but also into the nucleus and nucleolus of HeLa cells [23,24]. As an example, myelopeptide-4 has been shown to bind on the HL-60 cell surface, to penetrate into their cytoplasm, and finally to concentrate around the cell nucleus [23]. Recently it was shown that peptide EDR linked with ¡NG-containing de-oxyribooligonucleotides (preferably CAG-containing structures), making these sites unavailable to DNA methyltransferases, whereby the promoter was unmethylated [24,25].
Neurons cell cultures are an important models to study different diseases, allowing not only to simulate complex molecular mechanisms of the disease, but also to control various effects and search for potential therapeutic agents [26]. The correct development, functioning, viability of the striatal MSN and formation of functional dendritic spines require cortical neurons in cell culture. Therefore, in this study we used a physiological cell model of HD - mixed cortical-striatal neurons [27,28].
Our investigation was aimed to study the effect of the peptide EDR on the formation of MSN spines in the cortical-striatal neuronal cultures obtained from HD-bearing mice and to validate the hypothesis of DNA-peptide interactions underlying this effect.
Methods
Investigation of the KED peptide biological activity in the cortical-striatal cell culture
Cortical-striatal cell cultures were obtained from newborn wild-type (WT) (control) and transgenic FVB-Tg (YAC128)53Hay/J(YAC128) mice expressing human protein mHtt (The Jackson Laboratory, 004938, USA) [29]. These transgenic mice were used as a model to study HD. Cortical-striatal neuronal cultures of WT mice and YAC128 mice were divided each into 5 groups: 1-control (without adding peptides); 2-addition of peptide EDR (20 ng/ml), 3 - addition of peptide EDR (200 ng/ml); 4 – addition of peptide KED (Lys-Glu-Asp) 20 ng/ml; 5 - addition of peptide KED (200 ng/ml).
Initial sterile solutions of peptides EDR and KED (100 ng/ml) were diluted in Neurobasal medium (Invitrogen) till concentrations of 20 ng/ml and 200 ng/ml.
KED was known to reveal some angio-protective effects and was used as a control peptide. KED activated the process of cell renewal in organotypic and dissociated cell culture of vessels during its ageing, increasing the expression of Ki67 and reducing the synthesis of p53 protein and the synthesis of E-selectin - adhesion molecule involved in the formation of atherosclerotic plaques [30].
Genotyping
Heterozygous YAC128 males were crossed with WT females. The progeny was genotyped by polymerase chain reaction (PCR) with primers to exons 44 and 45 of the human huntingtin gene - RP - HD45 (5'-GCTCTGAAGGTATTCCTGGAT-3') and FP - HD44 (5'- TGTGCTCACGCTGTATGTGGA-3').
Neuronal cultures
Mixed cortical-striatal neurons in the cell culture were firstly used in our lab to study HD [27,28]. Cortex and striatum were obtained from WT and transgenic YAC128 (FVB-Tg (YAC128)53 Hay mice in the first day after birth. The isolated brain pieces were placed in a solution containing 1x isotonic saline Hank's buffer, (HBSS), 13.36 mM HEPES, 10 mM Na2CO3, 1x penicillin-streptomycin (PEST). Then the material was incubated with papain solution (Worthington) (37°C, 30 min), dissociated in the solution (10% fetal bovine serum (FBS), 0.5 mg/ml DNAase). Neurons were plated in 24 well plates containing 12 mM round Menzel cover slip (d0-1) precoated with 1% poly-D-lysine (Sigma) in Neurobasal medium (Invitrogen) supplemented with 2% B-27 (Gibco), 5% FBS and 1 mmol/L L- glutamine (Invitrogen). Cultures were incubated at 37°C and 5% CO2. To maintain cell cultures a solution, containing Neurobasal medium (Invitrogen) supplemented with 2% B-27 (Gibco) and 1 mmol/L L- glutamine (Invitrogen) at day in vitro (DIV) 7 and DIV14, was added. Peptide solutions were added to 2-5 groups of neuronal cell cultures 16 hours prior to fixation of neurons in the stated concentrations.
Immunostaining
Cells were fixed with solution (4% PFA + 4% sucrose in PBS) for 20 min at 4°C and permeabilized with 0.1% Triton-X-100. Fixed cell samples were blocked with 5% BSA, diluted in PBS (Blocking Solution), then immunostained with primary Anti-DARPP32 rabbit antibody (1:500) - striatal MSN marker, and Anti-MAP2 murine antibody (1:1000) - neuronal marker, followed by secondary antibodies (1:1000), conjugated with fluorophores (Alexa Fluor 488 and 594, Invitrogen). Then the coverslips were mounted in Mounting Media (Worrington).
Morphometric study and computer analysis of microscopy images
Z- Stack of optical section was captured with confocal microscope ThorLabs, X 100 oil -immersion objective lens (Olympus, UPlanSApo) and software ThorLabs (USA). Z interval was 0.2 μm. Each image was 1024 × 1024 pixels. We analyzed 7 neurons for each study group. Morphology analysis of dendritic spines was performed using the program Neuron Studio software package [31] following the procedure described in the article [32], allowing to automatically reconstruct a 3D image of spines and distribute them into three groups (mushroom, thin, hemp) according to specified parameters. To classify forms of spines of neurons in culture the following parameters were used: min stubby size - 50, non-stubby - 25, neck ratio - 1.4, thin ratio - 2.5, mushroom size - 0.5. The density distribution of spines was defined as an average number of spines 10 microns [31].
Statistical analysis
Statistical analysis of experimental data included calculation of the arithmetic mean, standard deviation and the confidence interval for each sample. Non-parametric U-Mann-Whitney test was used to compare the samples. In multiple comparisons, P<0.05 was recognized statistically significant.
Hypothesis testing of DNA-protein interactions in vitro and in silico
DNA-peptide interactions in a solution: Commercial calf thymus DNA (Sigma) with a molecular mass of 9 × 106 g mol-1 was used; molecular mass was calculated from the value of DNA intrinsic viscosity [η] in 0.15 ÃÂÂÂÂœ NaCl according to formula [η]=6.4 × 10-4 M 0.7 dl/g. DNA concentration in a stock solution was determined by the difference in the optical density of DNA solution at wavelengths of 270 nm and 290 nm after DNA hydrolysis in 6% HClO4 at 100°ÃƒÆ’¡ for 15 min. All reagents including EtBr and DAPI dyes were purchased from Sigma.
The complexes of DNA with peptide were prepared by mixing the equal volumes of DNA and peptide solutions in 5 mM NaCl. Before use, the resulting solutions were stored for 1 h at the ambient temperature (21°C). The triple complexes containing DNA, dye and peptide were also prepared by mixing the equal volumes of the solutions.
Gel electrophoresis experiments were carried out in the agarose gel (c=0.8%) with the use of 1TAE buffer. The experiments were performed at 60 V for 3 hours; then the gel was placed into the solution of ethidium bromide (EtBr) for 10 min and after that it was irradiated with UV lamp [33].
Absorption spectra were recorded with a SF-56 spectrophotometer (Russia) using 1 cm quartz cells. Luminescence was measured with a Hitachi-850 fluorescence spectrometer using 1 cm-thick quartz cuvette. Luminescence excitation and emission spectra were corrected for the spectral sensitivity of the instrument [34]. Relative viscosity of solutions ηr=η/η0 (where η is the solution viscosity, and η0 is the solvent viscosity) was measured with the Zimm-Crothers-type low-gradient rotation viscometer at flow gradients g = (0.1 ÷ 1) s-1 at 21°C. Then the reduced viscosity of DNA solutions with the same C(DNA) was calculated as (ηr-1)/C(DNA) [35].
Dynamic light scattering was studied using Photocor Complex (Russia), equipped with a semiconductor laser with a wavelength of 654 nm, and correlator Photocor-FC. The temperature required for DNA samples (21°C) was maintained thermostatically.
Molecular modelling in silico
System setup and solvation: The 10-bp DNA molecule was examined with the sequences of Poly(A), Poly(C), Poly(GC) and Poly(AT). EDR peptide was built in a left-handed stereo configuration. The three-dimensional DNA structures were modeled in the common B-form conformation. After having been constructed the molecules were protonated under conditions of pH=7 and T=300 Kelvin with optimization of their geometries in an all-atom force field Charmm27 [36]. Corresponding EDR-DNA complex was prepared. The simulations were run with the program Molecular Operating Environment 2014, (MOE 2014, Chemical Computing Group, Montreal, Canada) using the Charmm27 force field. The EDR-DNA complexes were solvated with the explicit TIP3P water model [37] extending to at least 10 Å beyond the DNA in each direction in a periodic cubic box (x=y=z=56 Å). Nineteen Na+ counterions were added to neutralize the system. The addition of the ions was carried out by random substitution of water oxygen atoms.
Docking. Modelling of the complexes of double-stranded DNA with EDR peptide was carried out using molecular docking method. This includes the measurement of bond energy and energetically favorable location of peptide in the DNA. For docking, DNA was chosen as a rigid body, peptide was chosen as a flexible body. A full DNA molecule was chosen as an active site. The generalized Born model [38] was used for implicit water simulation. London dG was used as the first and GBVI/WSA dG as the second scoring function for prediction of bond energy. Lower scores indicated more favorable poses, the unit for scoring function was kcal/mol. All molecular images were generated with the MOE 2014.
Results
Investigation of the biological activity of peptide KED in cortical-striatal cell culture
As previously shown [27,28] and afterwards confirmed in this study with a mixed DIV14 MSN cortical-striatal cell culture, an equal number of spines had been formed in a control (WT) and YAC128 cell cultures. It indicated the absence of negative impact of mHtt on spine morphology in the early stages of ontogenesis. At DIV21 elimination of spines occurred in YAC128 cultures (Figure 1). Moreover, many spines in YAC128 culture had filopodia-like morphology. This form is characteristic of immature or oppressed synapses. WT cultures retained the same density of spines at DIV21 as at 14DIV (Figure 1). These data evidenced degenerative processes in the aging MSN influenced by a mutant form of Htt and it was manifested in abuse of the formation and maintenance of spines. Figure 1 shows the WT and YAC128 cell cultures on 14th day in vitro (14DIV) and on 21st day in vitro (21DIV). We have found 14 spines in WT on 14DIV; 17 spines in WT 21DIV; 10 spines in YAC128 on 14; 6 spines in YAC128 on 21 DIV, 3 from which are mushroom-type spines. Figure 2 includes 9 spines, most of them are thin and stubby-type spines. Figure 2B shows 28 spines, most of them are functionally active mushroom-type spines. Figure 2C shows 15 various type of spines. In Figure 2D we have found 9 spines, most of them are thin and stubby-type spines.
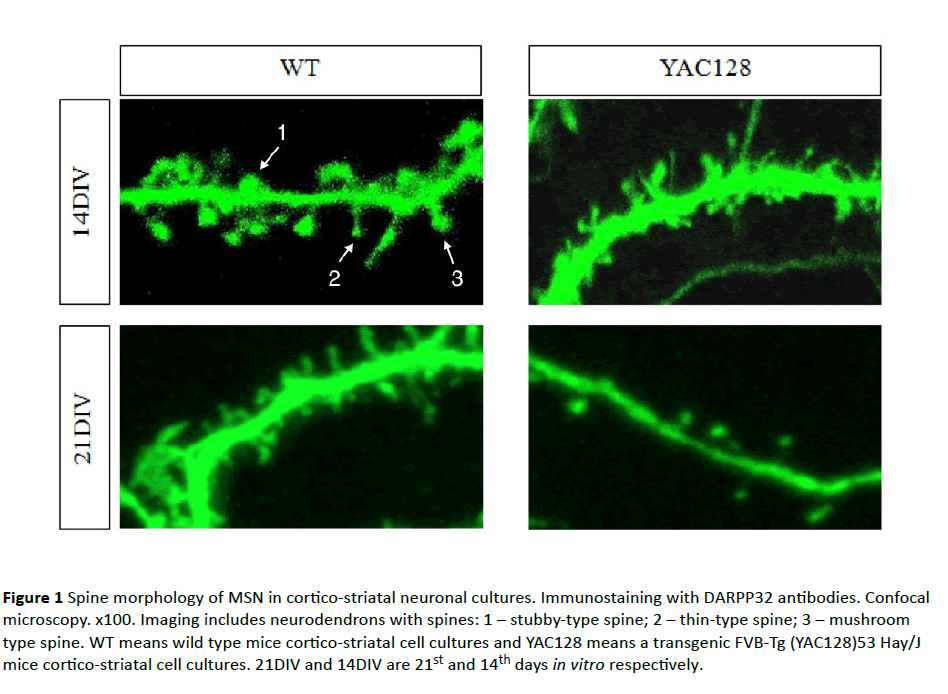
Figure 1 Spine morphology of MSN in cortico-striatal neuronal cultures. Immunostaining with DARPP32 antibodies. Confocal microscopy. x100. Imaging includes neurodendrons with spines: 1 – stubby-type spine; 2 – thin-type spine; 3 – mushroom type spine. WT means wild type mice cortico-striatal cell cultures and YAC128 means a transgenic FVB-Tg (YAC128)53 Hay/J mice cortico-striatal cell cultures. 21DIV and 14DIV are 21st and 14th days in vitro respectively.
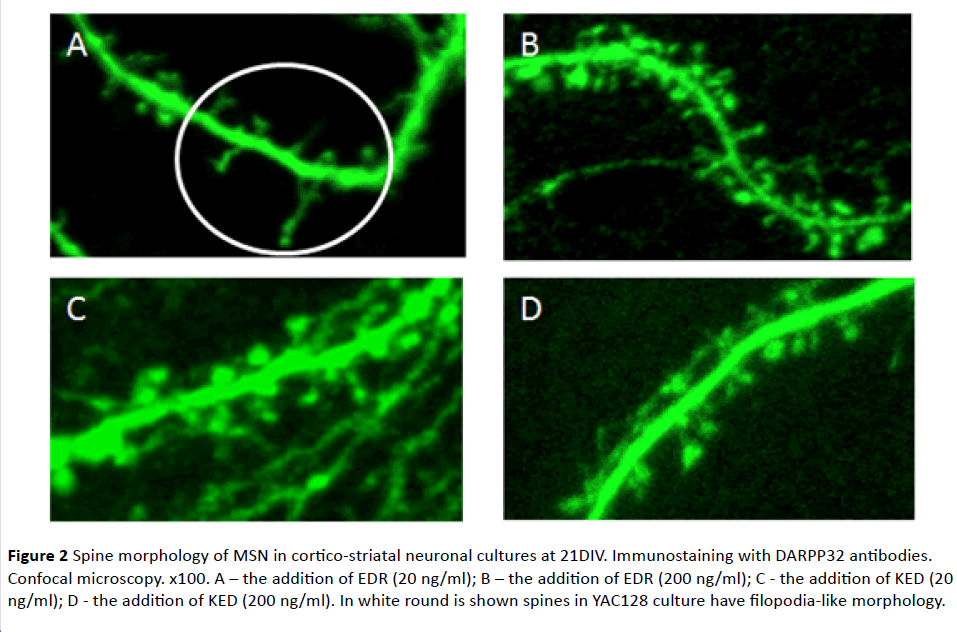
Figure 2 Spine morphology of MSN in cortico-striatal neuronal cultures at 21DIV. Immunostaining with DARPP32 antibodies. Confocal microscopy. x100. A – the addition of EDR (20 ng/ml); B – the addition of EDR (200 ng/ml); C - the addition of KED (20 ng/ml); D - the addition of KED (200 ng/ml). In white round is shown spines in YAC128 culture have filopodia-like morphology.
The effect of EDR and KED under study reference value of spine density was 8.4 ± 0.7 spines per 10 μm in WT cultures and 5.5 ± 0.4 spines per 10 μm in YAC128 cultures, which is by 34% lower than in WT (Figure 3).
Spine density increase to the reference value of WT mice was observed upon addition of EDR in concentration 200 ng/ml (Figure 2). Moreover, EDR affected the morphology of YAC128 neurons in concentration 20 ng/ml. On DIV21, spines acquired filopodia-like morphology: filopodia were more elongated (approximately twice or thrice, in individual cases up to 6 times) and branched compared with spines. However, there were no statistically significant differences in spine density between control and experimental samples. EDR in concentration 200 ng/ml had no effect on the morphology of neurons spines (Figure 2). KED in concentration 20 ng/ml had no effect on morphology and density of spines in YAC128 culture in any experiments. At the same time, in one of the experiments, KED at concentration 200 ng/ml promoted the increase of spines density up to 32% in culture YAC128 mice (Figures 2 and 3).
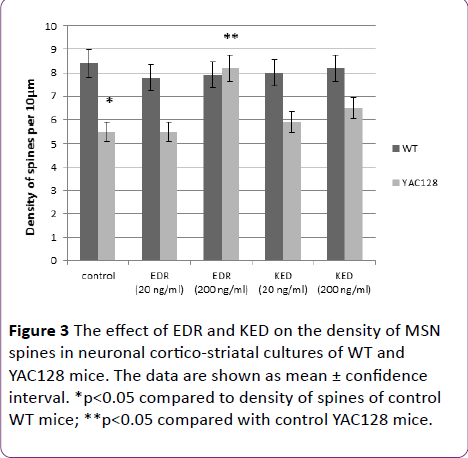
Figure 3 The effect of EDR and KED on the density of MSN spines in neuronal cortico-striatal cultures of WT and YAC128 mice. The data are shown as mean ± confidence interval. *p<0.05 compared to density of spines of control WT mice; **p<0.05 compared with control YAC128 mice.
Hypothesis testing of DNA-protein interactions in vitro and in silico
DNA-peptide interactions in a solution: EDR peptide contains three amino acids. In the aqueous solution at pH=7 peptide has three negative and two positive charges: N-terminal of peptide (pK=9.6), the guanidine group of arginine (pK=12.5), carboxyl groups of aspartic acid and glutamate (pK=3.9 and 4.2), and C-terminal (pK=2.17). The negative and positive charges of the peptide can be mutually attracted and these interactions affect the geometry of the peptide. DNA molecule in experimental conditions has negatively charged phosphate groups (pK=1.5). Experiments were carried out in 0.005 M NaCl solution at pH=7. High molecular calf thymus DNA in a solution had a conformation of a swollen statistical coil. EDR peptide does not absorb at wavelengths (λ) above 240 nm. Therefore, it was possible to monitor the state of DNA secondary structure during the interaction with EDR with absorption spectrum at λ>240 nm. The experiment proved that DNA absorption did not change in solutions with different EDR concentrations. We can conclude that the peptide does not affect spectral properties of DNA chromophores – base pairs. However, the peptide interacts with the DNA molecule, as it becomes clear from the following experiment (Figure 4). Different concentrations of the peptide were added to the DNA solution in 0.005 M NaCl at CDNA=constant before and after DNA binding with divalent ions or coordination compound cis-diammine, dichloroplatinum II, cis-DDP (anticancer drug cisplatin). The concentration of CuCl2 in DNA solution (10-5 M for all systems in Figure 4A) was chosen so that the vacant binding sites on DNA for copper ions were filled. It is known that Cu2+ ions interact with DNA phosphates and N7 Guanine in a major groove and induce typical changes in the DNA absorption spectrum. Such binding causes the bathochromic shift of the DNA absorption band [39], which is observed also in all DNA solutions containing copper ions and peptides at r<3 (Figure 4A). We could detect Cu2+ interaction with DNA in solutions with the exception of solution with r=3 where the peptide binding prevailed. It is believed that at r=3 the peptide prevents the copper ions to be located in the major groove of DNA near N7 Guanine (the typical binding sites for Cu2+ on DNA). Thus, we assume that at high concentrations of the peptide in DNA solution, EDR overlaps the major groove and blocks Cu2+ binding to N7 Guanine, if we exclude possible binding of the peptide with copper ions. Indeed, the experimental data indicate that the addition of the peptide and Cu2+ in different sequences into DNA solution leads to the same result.
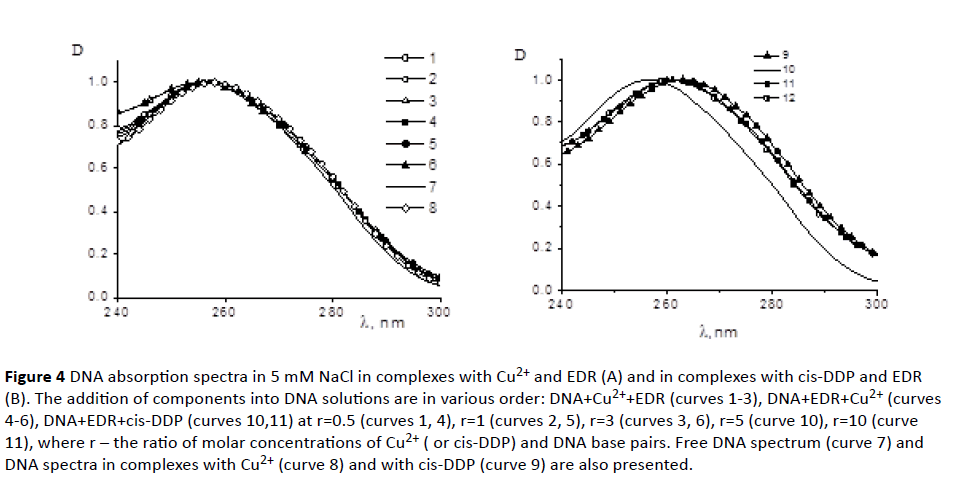
Figure 4 DNA absorption spectra in 5 mM NaCl in complexes with Cu2+ and EDR (A) and in complexes with cis-DDP and EDR (B). The addition of components into DNA solutions are in various order: DNA+Cu2++EDR (curves 1-3), DNA+EDR+Cu2+ (curves 4-6), DNA+EDR+cis-DDP (curves 10,11) at r=0.5 (curves 1, 4), r=1 (curves 2, 5), r=3 (curves 3, 6), r=5 (curve 10), r=10 (curve 11), where r – the ratio of molar concentrations of Cu2+ ( or cis-DDP) and DNA base pairs. Free DNA spectrum (curve 7) and DNA spectra in complexes with Cu2+ (curve 8) and with cis-DDP (curve 9) are also presented.
To clarify this issue, we used the compound cis-DDP (Figure 4B) which formed a coordination bond with the nitrogen bases of DNA (primarily with N7 guanine). Such linkages were not broken by the addition of the peptide. The experiment showed that the binding of cis-DDP with DNA prevailed in all systems, although peptide had a slight effect on the spectral properties of a DNA molecule. The latter could point out that peptide also interacted with DNA. DNA CD spectra in the presence of peptides did not change. These data confirmed the absence of peptide influence on the DNA secondary structure. However, the peptide may change base stacking and weaken the hydrogen bonds between complementary bases (without noticeable influence on DNA CD spectrum). This assumption has been confirmed by the study of DNA melting in the complexes with EDR (Figure 5A). As seen from the experimental results, the used concentration of the peptide slightly destabilizes DNA. A small decrease in the melting temperature of the DNA in the complexes with the peptide indicate that at r=3 the DNA destabilization was already observed.
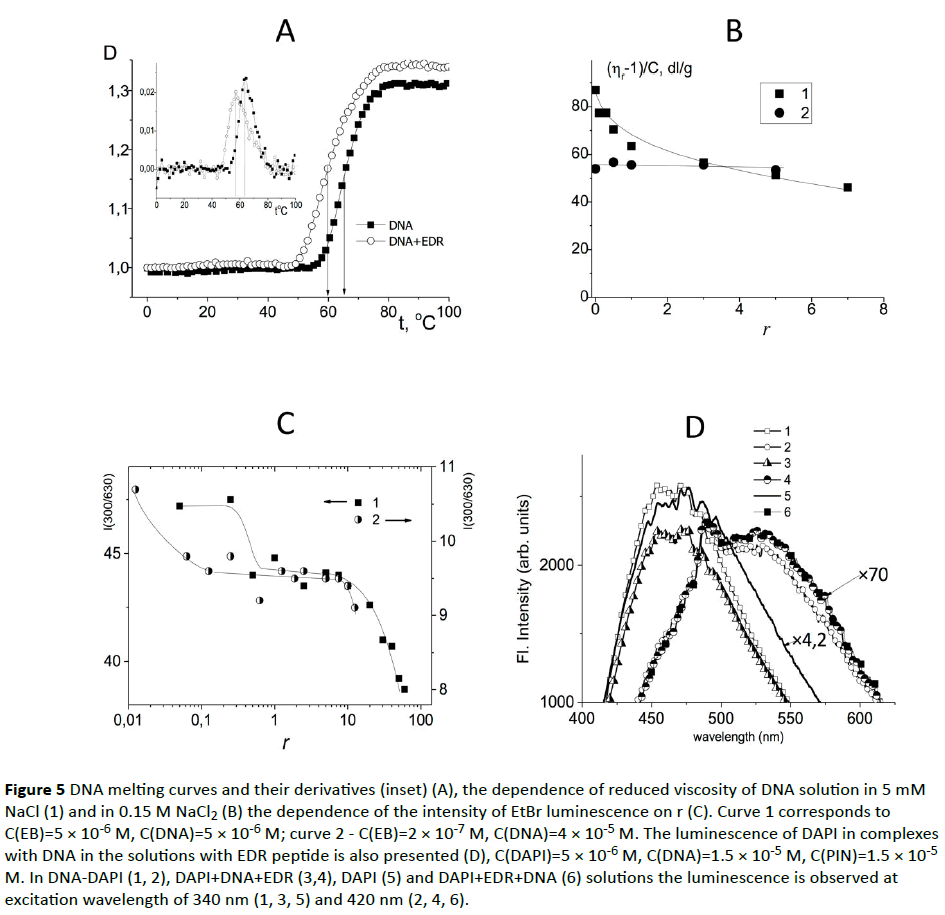
Figure 5 DNA melting curves and their derivatives (inset) (A), the dependence of reduced viscosity of DNA solution in 5 mM NaCl (1) and in 0.15 M NaCl2 (B) the dependence of the intensity of EtBr luminescence on r (C). Curve 1 corresponds to C(EB)=5 × 10-6 M, C(DNA)=5 × 10-6 M; curve 2 - C(EB)=2 × 10-7 M, C(DNA)=4 × 10-5 M. The luminescence of DAPI in complexes with DNA in the solutions with EDR peptide is also presented (D), C(DAPI)=5 × 10-6 M, C(DNA)=1.5 × 10-5 M, C(PIN)=1.5 × 10-5 M. In DNA-DAPI (1, 2), DAPI+DNA+EDR (3,4), DAPI (5) and DAPI+EDR+DNA (6) solutions the luminescence is observed at excitation wavelength of 340 nm (1, 3, 5) and 420 nm (2, 4, 6).
It is reasonable to consider a possible peptide influence on the DNA tertiary structure. Figure 5B shows the dependence of the reduced viscosity of DNA solutions at C(DNA)=constant on the relative peptide/DNA concentration in solutions with low (5 mM) and physiological (15 M) NaCl concentrations. According to the experimental data, the presence of peptide in the DNA solution of physiological ionic strength (0.15 M NaCl) does not influence the reduced viscosity of DNA solution. This indicates that the volume of DNA coil does not change. Opposite, at low NaCl concentration (0.005 M) there is a gradual decrease in the viscosity with the rise of r, which reflects the shrinkage of DNA molecular coil due to interaction with peptide. At r>6 the viscosity reaches the value which is less than observed for the same DNA concentration in 0.15 M NaCl and approximately equal to that observed in 1 M NaCl at complete suppression of the polyelectrolyte swelling. Hence it can be concluded that the binding of the peptide influences the volume effects in DNA solutions. Presumably the binding of the peptide to the DNA molecule is carried out due to the winning in the entropy of systems as a result of the release of water molecules from the hydration shells of the DNA and the peptide. At high NaCl concentrations DNA hydration sell can be destabilized by ion implantation, as a consequence of either binding does not occur, or it does not lead to a drop in the volume of the coil.
In addition, the electrostatic interactions can play an important role in the formation of DNA-peptide complexes. Indeed, in 0.15 M NaCl the shielding of DNA phosphate groups prevented the interaction of the positively charged groups of peptide with DNA. In 0.005 M NaCl the viscosimetric data clearly demonstrated that the peptide interacted with DNA. This interaction was accompanied by decreasing in DNA polyelectrolyte swelling (thermodynamic volume effects could not produce the observed DNA shrinkage). In contrast, the results received by electrophoresis didn’t demonstrate any difference in electrophoretic mobility of DNA Indeed, the binding of peptides to DNA could be destroyed during the experiment. Nevertheless, not only viscometry, but also the data of dynamic light scattering experiment evidence the formation of DNA complexes with peptides which affect the molecular size of the DNA coil (Table 1). Let us consider the effect of peptide presence in DNA solution on the luminescence of EtBr and DAPI dyes associated with DNA (Figure 5C). At certain concentration of the peptide in DNA solution the quenching of the ethidium bromide (EtBr) fluorescence was observed. We used two dye concentrations with their relation to the concentration of DNA base pairs z=0.5 and z=0.005 (z – is the ratio of the molar concentration of the dye and the DNA base pairs). At both concentrations of EtBr the luminescence quenching was observed at a sufficiently low concentration of peptide r>0.2. A sharp quenching of the luminescence recorded at r>10 (Figure 6A) was observed. At r=0.005 the decrease in EtBr luminescence began at substantially lower r (peptide-to -DNA concentration ratio). As it is known, the EtBr intercalates between the DNA bases, and at high dye concentrations the external binding with DNA phosphates is also realized.
| C(DNA) |
r, bp |
RH, nm |
| 0.001% |
0 |
740 ± 180 |
| 0.001% |
1 |
700 ± 150 |
| 0.001% |
3 |
560 ± 120 |
| 0.001% |
10 |
310 ± 70 |
C(DNA) - Concentrations of DNA; r=C(EDR )/C(DNA); bp – base pair
Table 1 Values of hydrodynamic radius of DNA and DNApeptide complexes in 0.005 M NaCl obtained with the dynamic light scattering method.
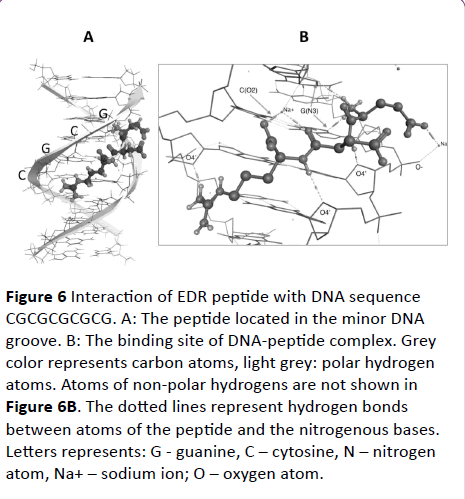
Figure 6 Interaction of EDR peptide with DNA sequence CGCGCGCGCG. A: The peptide located in the minor DNA groove. B: The binding site of DNA-peptide complex. Grey color represents carbon atoms, light grey: polar hydrogen atoms. Atoms of non-polar hydrogens are not shown in Figure 6B. The dotted lines represent hydrogen bonds between atoms of the peptide and the nitrogenous bases. Letters represents: G - guanine, C – cytosine, N – nitrogen atom, Na+ – sodium ion; O – oxygen atom.
Another dye DAPI binds to DNA in two ways (Figure 5D). At low DAPI concentration in DNA solution dye is located in the minor groove of DNA, forming hydrogen bonds with partial intercalation between DNA bases. The luminescence band which reflects this type of binding has a maximum 450 nm (λex=340 nm). The second type of binding is the external electrostatic DAPI binding with the phosphate groups of DNA. This type of binding is expressed in the luminescence band with a maximum of 530 nm (λex=420 nm). The luminescence quantum yield for the second type binding is much less. Solutions containing a dye, a peptide and DNA molecules were prepared at z=C(DAPI)/C(bp)=0.3. At this value of z both types of DAPI binding to DNA could be observed with the luminescence. Indeed, we were able to see the contribution of the external binding: at 420 nm excitation the luminescence spectra was observed with maximum at 530 nm for the electrostatic DNA-DAPI complexes in 0.005 M NaCl. The excitation spectra did not change. The excitation at 340 nm caused the luminescence of DAPI with the band maximum at 450 nm to 475 nm (strong binding in DNA minor groove). The addition of the peptide (Figure 5D) to DNA-DAPI complexes slightly reduced the intensity of DAPI luminescence band which related to a dye binding in the minor groove, and had little influence on the second type of binding (the external electrostatic binding). The peptide effect on the DAPI luminescence in a solution without DNA was not registered (the intensity and shape of the excitation and emission spectra were not changed).The analysis of the data obtained brings to the conclusion that the addition of EDR into DAPI-DNA solution at used concentrations influenced the luminescence of DAPI, participating in the formation of a strong binding (in the minor groove) and slightly increased the luminescence of external binding. The excitation spectra were not changed. The above data suggest that the peptide interacts with the DNA molecule in a solution with reducing dye luminescence, which is localized in the minor groove of the DNA, while EDR has a slight effect on externally connected dye. In this way, the localization of peptide on DNA can be changed due to the presence of other binding ligands (DAPI or EtBr).
Molecular modelling in silico: Overview of computational model. The analysis of spectral data (UV absorption and CD spectra) indicates that the EDR peptide interacts with DNA in water-salt solution via nitrogen bases. The present simulations of DNA bound to EDR peptide allows to find out specific interactions between DNA double helix and peptide in solution, which is conformed to experimental data. Recent study shows that EDR predominantly quenched fluorescence of oligo(dT) and oligo(dGC) in experiments with single- and double- stranded fluorescence-labeled deoxyribooligonucleotides [23]. Therefore, selective peptide binding with CG promoter sites may protect them from the action of respective DNA-methyltransferases which may be crucial for activation of most genes.
We examined DNA complexed to EDR peptide in aqueous solution with conterion addition in Charmm27 force field using methods of molecular minimization and docking (see Materials and Methods). Docking experiments were performed using default parameters with MOE. From analysis docked structures, we concluded that the EDR peptide is stabilized by one or more H-bonds with the DNA bases. In all cases, peptide binds DNA along the minor groove. It was found that the energy pose of DNA interaction with EDR for oligo(dCG) was lower than for other oligonucleotides (dG=-7.5 kcal/mol) that points on the formation of the energetically effective complex. Thus the peptide predominantly binds with the oligonucleotide that has more cytosine-guanine repeats.
Figure 6 shows localization of peptide in proposed binding site of DNA. The edges of the bps, which form the bottom of the grooves, contain nitrogen and oxygen atoms available for contacting protein side chains via hydrogen bonds. We investigated the formation of hydrogen bonds between peptide and DNA fragment. The hydrogen bonds formed the basis of sequence-specic recognition of DNA by proteins. The binding site and energy of interaction was examined using docking method. Peptide at DNA interfaces could interact with DNA directly through hydrogen bonds and van der Waals contacts, indirectly through water-mediated hydrogen bonds or through Na+ ions (Figure 6B).
Discussion
It was established in experiments on cell cultures that the peptide EDR possessed neuroprotective properties which promoted restoration of striatal neuron spine’s morphology in mice of HD model. It is well known that a compendial drug of heptapeptide (GP, Met-Glu-His-Phe-Pro-Gly-Pro), manifests anti-inflammatory, antioxidant, nootropic action [40], similar to peptide EDR. In this case such effects are typical not only for the heptapeptide, but also for its metabolites – tripeptides [41]. For Semax in the study on patients of elderly and old age with initial stages of HD it has been shown the improvement of the quality of life, improving of motor and social activity and memory after intranasal administration of the drug. However, these data were obtained using questionnaires MMSE, ADAS-cog, GGI, IADL and could not serve as an objective evidence for drug efficacy. Objective improvement of brain functions in patients with HD under the influence of Semax consisted in normalization of the EEG rhythms [42]. The mechanism of Semax neuroprotective effect and its impact on the morphology of neurons in HD have not been studied. EDR peptide possibly has radio-protective properties similar to those of hexapeptide Semax. In view of its ability to restore the morphology of spines striatum neurons, it can potentially be a promising drug for the treatment of HD.
Previous investigations have shown that short peptides especially those which include positively charged lysine or arginine may penetrate into the cell and nucleus and probably bind to the DNA [24,25]. So, peptide KEDW (Lys-Glu-Asp-Trp-NH2) is able to reduce the blood glucose level in rats with streptozotocin- and alloxan-induced diabetes mellitus. KEDW peptide increased the expression of PDX1, NGN3, PAX6, FOXA2, NKX2-2, NKX6.1, and PAX4 genes but decreased MNX1 and HOXA3 gene expression when added to pancreatic cell culture [42]. KEDW peptide caused an increase in expression of PDX1, NGN3, PAX6, FOXA2, NKX2-2, NKX6.1, and PAX4 proteins without affecting synthesis of MNX1 and HOXA3 when added to pancreatic cell culture. Results obtained through physical methods (UV–visible absorption, circular dichroism) and molecular modelling methods suggest that the peptide binds to DNA along the major groove. Experimental and theoretical data provided a 3D model of a stable DNA-peptide complex [43]. Another peptide, AEDL, regulates the levels of Ki67, Mcl-1, p53, CD79, NOS-3 proteins in cell cultures of human bronchial epithelium in various passages. The strongest activating effect of peptide ADEL on bronchial epithelial cell proliferation through Ki67 and Mcl-1 was observed in old cell cultures. ADEL regulateed the expression of genes involved in bronchial epithelium differentiation: NKX2-1, SCGB1A1, SCGB3A2, FOXA1, and FOXA2. ADEL also activateed several genes, which reduced expression correlated with pathological lung development: MUC4, MUC5AC, and SFTPA1. Spectrophotometry, viscometry, and circular dichroism showed ADEL–DNA interaction, with a binding region in the major groove (N7 guanine) [44].
In this work the data collection of DNA-peptide interactions in solution showed that the character of peptide-DNA binding depended on peptide concentration in solution. With a sufficiently large concentration of peptide it inhibited the binding of the compounds on the major groove of DNA and had little effect on the binding components on the minor groove. Furthermore, the presence of peptide extinguishes the ethidium bromide luminescence in DNA solution. It can be connected, for example, with gradual formation of a double helix peptide coverage and local change of permittivity of the medium near the macromolecule.
The present simulations of DNA binding to the EDR peptide allow the understanding of short peptide properties in the cell. Combination of the data obtained from the present analysis of EDR–DNA complex and from experimental studies suggests a recognition mechanism by peptides. Based on the established biological activity of the peptide and our experimental data we can assume that small peptides (di-, tri- and tetrapeptides) revealed capability to interact with DNA in the area of specific binding site on the promoter segment of genes.
Discovery of the phenomenon of peptide activation of gene transcription points out the natural mechanism of organism to maintain physiologic functions, which is based on the interaction of the DNA and regulatory peptides. This process is fundamental for the development and functioning of the living substance. Thus, the proposed models may serve for studying mechanisms of biological activity of short peptides and quantitative assessment of their expected activity.
Conclusion
This work has demonstrated that the peptide EDR normalized spines morphology of neurons in a mouse model of HD and interacted with DNA in a solution. This effect points to the ability of EDR peptide to regulate homeostasis in neurons. According to the previous data and present study we propose that EDR peptide penetrates into neurons and regulates a number of dendritic spines through binding with nucleotides.
Thus, the peptide EDR as a representative of a pool of regulatory biologically active peptides, holds promise as one of neuroprotective agents that are encouraging for further study as a compound effective for the treatment of HD.
Funding
This work was supported by the state grant 17.1360.2014 (results showed on Figure 1), by the Russian Science Foundation Grant 14-25-00024 (results showed on Figures 2 and 3).
Competing Interests
The authors declare that they have no competing interests.
18199
References
- MacDonald ME, Ambrose CM, Duyao MP, Myers RH, Lin C, et al. (1993) A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell 72: 971-983.
- Kremer B, Goldberg P, Andrew SE, Theilmann J, Telenius H, et al. (1994) A worldwide study of the Huntington’s disease mutation. The sensitivity and specificity of measuring CAG repeats. N Engl J Med 330: 1401–1406.
- Rubinsztein DC, Leggo J, Coles R, Almqvist E, Biancalana V, et al. (1996) Phenotypic characterization of individuals with 30-40 CAG repeats in the Huntington’s disease (HD) gene reveals HD cases with 36 repeats and apparently normal elderly individuals with 36-39 repeats. Am J Hum Genet 59: 16–22.
- Quarrell OWJ, Rigby AS, Barron L, Crow Y, Dalton A, et al. (2007) Reduced penetrance alleles for Huntington’s disease: a multi-centre direct observational study. J Med Genet 44: e68.
- Lu B, Palacino J (2013) A novel human embryonic stem cell-derived Huntington’s disease neuronal model exhibits mutant huntingtin (mHTT) aggregates and soluble mHTT-dependent neurodegeneration. FASEB J Off Publ Fed Am Soc Exp Biol 27: 1820-1829.
- Harjes P, Wanker EE (2003) The hunt for huntingtin function: Interaction partners tell many different stories. Trends Biochem Sci 28: 425-433.
- Ross CA (2002) Polyglutamine pathogenesis: Emergence of unifying mechanisms for Huntington’s disease and related disorders. Neuron 35: 819-822.
- Sugars KL, Rubinsztein DC (2003) Transcriptional abnormalities in Huntington disease. Trends Genet 19: 233-238.
- Tobin AJ, Signer ER (2000) Huntington’s disease: The challenge for cell biologists. Trends Cell Biol 10: 531-536.
- Tang TS, Tu H, Chan EYW, Maximov A, Wang Z, et al. (2003) Huntingtin and huntingtin-associated protein 1 influence neuronal calcium signaling mediated by inositol-(1,4,5) triphosphate receptor type 1. Neuron 39: 227-239.
- Tang TS, Chen X, Liu J, Bezprozvanny I (2007) Dopaminergic signaling and striatal neurodegeneration in Huntington’s disease. J Neurosci 27: 7899-7910.
- Tang TS, Guo C, Wang H, Chen X, Bezprozvanny I (2009) Neuroprotective effects of inositol 1,4,5-trisphosphate receptor C-terminal fragment in a Huntington’s disease mouse model. J Neurosci 29: 1257-1266.
- Wu J, Tang T, Bezprozvanny I (2006) Evaluation of clinically relevant glutamate pathway inhibitors in in vitro model of Huntington’s disease. Neurosci Lett 407: 219-223.
- Zhang H, Li Q, Graham RK, Slow E, Hayden MR, et al. (2008) Full length mutant huntingtin is required for altered Ca2+ signaling and apoptosis of striatal neurons in the YAC mouse model of Huntington’s disease. Neurobiol Dis 31: 80-88.
- Arutjunyan A, Kozina L, Stvolinskiy S, Bulygina Y, Mashkina A, et al. (2012) Pinealon protects the rat offspring from prenatal hyperhomocysteinemia. Int J Clin Exp Med 5: 179-185.
- Mendzheritskii AM, Karantysh GV, Abramchuk VA, Ryzhak GA (2014) Effect of peptide geroprotectors on navigation learning in rats of different ages and caspase-3 systems in their brain structures. Adv Gerontol 4: 37-41.
- Khavinson V, Ribakova Y, Kulebiakin K, Vladychenskaya E, Kozina L, et al. (2011) Pinealon increases cell viability by suppression of free radical levels and activating proliferative processes. Rejuvenation Res 14: 535-541.
- Trabulo S, Cardoso AL, Mano M, De Lima MCP (2010) Cell-penetrating peptides—Mechanisms of cellular uptake and generation of delivery systems. Pharmaceuticals 3: 961-993.
- Takeshima K, Chikushi A, Lee KK, Yonehara S, Matsuzaki K (2003) Translocation of analogues of the antimicrobial peptides magainin and buforin across human cell membranes. J Biol Chem 278: 1310-1315.
- Kubo T, Yokoyama K, Ueki R, Abe S, Goto K, et al. (2000) Structure and affinity of DNA binding peptides. Nucleic Acids Symp Ser 44: 49–50.
- Solovyev AY, Tarnovskaya SI, Chernova IA, Shataeva LK, Skorik YA (2015) The interaction of amino acids, peptides, and proteins with DNA. Int J Biol Macromol 78: 39-45.
- Fedoreyeva LI, Kireev II, Khavinson VK, Vanyushin BF (2011) Penetration of short fluorescence labeled peptides into the nucleus in HeLa cells and in vitro specific interaction of the peptides with deoxyribooligonucleotides and DNA. Biochem Moscow 76: 1210-1219.
- Guryanov SA, Kirilina EA, Khaidukov SV, Suvorov NI, Molotkovskaya IM, et al. (2006) Fluorescently labeled differentiating myelopeptide-4: Specific binding to and penetration into target cells. Russ J Bioorganic Chem 32: 517-520.
- Khavinson VK, Soloveva Y, Tarnovskaya SI, Linkova NS (2013) Mechanism of biological activity of short peptides: Cell penetration and epigenetic regulation of gene expression. Biol Bull Rev 3: 451-455.
- Ariano MA, Aronin N, Difiglia M, Tagle DA, Sibley DR, et al. (2002) Striatal neurochemical changes in transgenic models of Huntington’s disease. J Neurosci Res 68: 716-729.
- Artamonov DN, Korzhova VV, Wu J, Rybalchenko PD, Im K, et al. (2013) Characterization of synaptic dysfunction in an in vitro corticostriatal model system of Huntington’s disease. Biochem Suppl Ser A Membr Cell Biol 7: 192-202.
- Wu J, Ryskamp DA, Liang X, Egorova P, Zakharova O, et al. (2016) Enhanced store-operated calcium entry leads to striatal synaptic loss in a Huntington’s disease mouse model. J Neurosci 36: 125-141.
- Slow EJ, Van Raamsdonk J, Rogers D, Coleman SH, Graham RK, et al. (2003) Selective striatal neuronal loss in a YAC128 mouse model of Huntington’s disease. Hum Mol Genet 12: 1555-1567.
- Khavinson VK, Tarnovskaia SI, Linkova NS, Guton EO, Elashkina EV (2014) Epigenetic aspects of peptidergic regulation of vascular endothelial cell proliferation during aging. Adv Gerontol 27: 108-114.
- Rodriguez A, Ehlenberger DB, Dickstein DL, Hof PR, Wearne SL (2008) Automated three-dimensional detection and shape classification of dendritic spines from fluorescence microscopy images. PLoS One 3: e1997.
- Sun S, Zhang H, Liu J, Popugaeva E, Xu NJ, et al. (2014) Reduced synaptic STIM2 expression and impaired store-operated calcium entry cause destabilization of mature spines in mutant presenilin mice. Neuron 82: 79–93.
- Pellegrino T, Sperling RA, Alivisatos AP, Parak WJ (2007) Gel electrophoresis of gold-DNA nanoconjugates. J Biomed Biotechnol 2007: 26796.
- Qiao C, Bi S, Sun Y, Song D, Zhang H, et al. (2008) Study of interactions of anthraquinones with DNA using ethidium bromide as a fluorescence probe. Spectrochim Acta A Mol Biomol Spectrosc 70: 136-143.
- Frisman EV, Shchagina LV, Vorobev VI (1965) A glass rotating viscometer. Biorheology 2: 189-194.
- Mackerell ADJ, Feig M, Brooks CL 3rd (2004) Extending the treatment of backbone energetics in protein force fields: Limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comput Chem 25: 1400-1415.
- Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79: 926-935.
- Onufriev A, Case DA, Bashford D (2002) Effective Born radii in the generalized Born approximation: the importance of being perfect. J Comput Chem 23: 1297-1304.
- Kasianenko NA, Diakonova NE, Frisman EV (1989) A study of the molecular mechanism of DNA interaction with divalent metal ions. Mol Biol 23: 975-982.
- Yatsenko KA, Glazova NY, Inozemtseva LS, Andreeva LA, Kamensky AA, et al. (2013) Heptapeptide semax attenuates the effects of chronic unpredictable stress in rats. Dokl Biol Sci 453: 353-357.
- Iasnetsov VV, Chertorizhskii EA, Belyi PA, Bespalova ZD, Ovchinnikov MV, et al. (2015) Study of neuroprotective, antihypoxic and antiamnesic effects of new mixture of tripeptides. Eksp Klin Farmakol 78: 3-8.
- Kaplan AYA, Kochetova AG, Nezavibathko VN, Rjasina TV, Ashmarin IP (1996) Synthetic ACTH analogue semax displays nootropic-like activity in humans. Neurosci Res Commun 19: 115-123.
- Khavinson VK, Tendler SM, Kasyanenko NA, Tarnovskaya SI, Linkova NS, et al. (2015) Tetrapeptide KEDW interacts with DNA and regulates gene expression. Am J Biomed Sci 7: 156-169.
- Khavinson VK, Tendler SM, Vanyushin BF, Kasyanenko NA, Kvetnoy IM, et al. (2014) Peptide regulation of gene expression and protein synthesis in bronchial epithelium. Lung 192: 781-791.