Keywords
CTX-M; Escherichia coli; Animal origin; Tunisia
Introduction
E. coli is a common intestinal microorganism of humans and animals. It also represents an important pathogen causing urinary tract and gastrointestinal infections and septicaemia [1]. Over the past few years, resistance to antimicrobial agents has increased among E. coli from both human and animal sources [2]. Initially, resistance was described to particular agents, such as ampicillin, trimethoprim, sulphur-based antimicrobials or tetracyclines [3]. Recently, the resistance has broadened with the emergence of broad resistance to large families of antimicrobial agents among human and veterinary isolates. This resistance seems to be in relation to their wide clinical use of these agents. The mechanisms of resistance to oxyimino-cephalosporins in E. coli isolates from animals and food products by the production of extended-spectrum β-lactamases (ESBLs) have become prominent [4-6]. Recent reports describing the detection of ESBLs in bacterial isolates from animals have raised concern regarding the transmission of ESBL genes between human and animal isolates [2]. E. coli strains carrying AmpC-β-lactamases such as CMY-type lactamase have already been reported in healthy and sick animals and food-producing animals [7-9]. In addition to the resulting resistance to most β-lactam antibiotics, ESBL or plasmidic AmpC-β-lactamases producers are also frequently resistant to aminoglycosides and fluoroquinolones. The rate of resistance to quinolones and fluoroquinolones among E. coli isolates of animal origin has been increasingly reported [10,11]. The prevalence of antibiotic resistance in E. coli of animal origin has increased due to the inclusion of different antimicrobial resistance genes on mobile elements such as plasmids, transposons and integrons that facilitate the rapid dissemination of these genes among bacteria [12]. Commensal E. coli seems to be the major reservoir of resistance gene since they have been implicated in the transmission of genetic traits from one bacterium to another [13]. The impact of animal-derived broad-spectrum-β-lactamases (ESBL)-producing E. coli on public health has drawn considerable attention worldwide [2,13]. The high level of increase of antibiotic resistance in E. coli of animal origin may be enhanced by the use of antimicrobials for preventive purposes. This practice could have been followed in Tunisia and have been probably applied to cover up the failures in farm management. Epidemiological studies on the antibiotic resistance in E. coli in Tunisia are relatively new; the first published paper appeared in 2007. Since then, more data have been made available and the real situation of antibiotic resistance in E. coli from animal origin is very alarming. In the present review, we focus on the analyses of ESBL-producing E. coli of animal origin in Tunisia and the genetic support of this resistance.
ESBL encoding genes in Tunisia
A chronologically listed summary of β-lactamases in E. coli from food-producing animals in Tunisia is given in Table 1. The detection of ESBL-producing Enterobacteriaceae that derivate from genes originally encoded for TEM-1, TEM-2 or SHV-1 has been increasingly reported in the last few years in Tunisia [14- 16]. However, interestingly the epidemiology of ESBLs in E. coli has changed radically: novel ESBLs such as CTX enzymes are predominant, replacing the classical TEM and SHV-type ESBL in many countries as well as in Tunisia [16].
| References |
Year of isolation |
ESBL and AmpC detected |
Origin |
| Jouini et al. 2007 |
2006 |
CTX-M-1 |
Beef, turkey |
| |
|
CTX-M-1 + TEM-1 |
Beef |
| |
|
CTX-M-14+ TEM-1 |
Chicken |
| |
|
CTX-M-8 |
Chicken |
| |
|
SHV-5 |
Chicken |
| |
|
TEM-1 |
Turkey |
| Ben Slama et al.2010 |
2007 |
CTX-M-1 |
Chicken, sheep, beef |
| |
|
CTX-M-1 + TEM-20 |
Sheep |
| |
|
CTX-M-1 + TEM-1b |
Chicken |
| |
|
CMY-2 |
Chicken |
| Mnif et al. 2012 |
2010 |
CTX-M-1, TEM-1, CTX-M-15, CMY-2 |
fecal samples of healthy chickens |
| Ben Sallem et al.2012 |
2011 |
CTX-M-1, TEM-135, CMY-2, TEM-1b |
fecal samples of healthy chickens |
| |
|
CTX-M-1 |
fecal samples of healthy dromedary |
| Ben Sallem et al.2013 |
2010 |
CTX-M-1, TEM-1c, TEM-135, TEM-1b, CMY-2 |
fecal samples of healthy pets |
| Grami et al.2013 |
2011-2012 |
CTX-M-1 |
Fecal samples of chicken and healthy pets |
| |
|
CTX-M-15 |
Fecal sample of dog |
| |
|
CTX-M-9 |
Fecal sample of chicken |
| Grami et al. 2014 |
2010-2011 |
CTX-M-15 |
Diseased cattle milk |
| Kilani et al 2015 |
2013-2014 |
CTX-M-15, TEM-1 |
Healthy chicken |
Table 1: The occurrence of β-lactamases reported in E. coli of animal origin in Tunisia.
In Tunisia and in other African countries, the first CTX-M ESBLtype was detected in 2006 in samples of food origin plated onto selective medium with cefotaxime [17]. The ESBLs detected were blaCTX-M-1, blaCTX-M-14, blaCTX-M-8 and blaSHV-5, this was the first report of CTX-M-8 occurrence in Tunisia. In addition, the aforementioned report described the first detection of blaCTX-M-14 and blaSHV-5 genes in 2/11 and 1/11 E. coli isolates from faecal samples of chicken. In Europe, Danish and British ESBL-producing E. coli data from 2012 revealed the presence of blaCTX-M-14 gene respectively in pigs and in poultry; however blaSHV-5 gene was detected in Spain from pigs (Blanc et al. 2006) [18,19]. Chinese data demonstrated the occurrence of CTX-M-14 in combination with CTX-M-1 type ESBLs recovered from healthy and sick pets; SHV-5 was also isolated from livestock animals in Japan [1,20].
Since the number of food samples of animal origin used by Jouini et al. was small (38 samples), similar studies have been conducted by Ben Slama et al. in the same laboratory to characterize ESBLproducing E. coli recovered from 79 food samples of animal origin during 2007 [17,12]. Only the CTX-M-1 type ESBL was detected in this study. Since 2007, CTX-M-1 has been continuing to appear in Tunisia. Indeed, Mnif et al. have reported a high frequency of cefotaxime-resistant E. coli isolates from healthy chicken faecal samples (67 isolates, 42%, recovered from 24 of 36 Tunisian farms located in six different governorates of Tunisia) [21]. Again, all ESBLs (43 strains) belonged to CTX-M group 1: 39 CTX-M-1 and 4 CTX-M-15, and the AmpC phenotype (observed in 24 strains) harbored the blaCMY-2 gene (Figure 1). This was the first report on CTX-M β-lactamases in live broiler chickens and the first report of blaCTX-M-15 gene in E. coli strains from animal origin in Tunisia. CTX-M-15 has been identified in food-producing animals for the first time in France in one isolate recovered from poultry, then in Denmark from pigs and in Argentina and Great Britain from poultry [18,19,22,23]. Recently, a novel investigation reported the detection of CTX-M-15 type ESBL in diseased cattle from milk sample for the first time in Tunisia and even Africa [24].
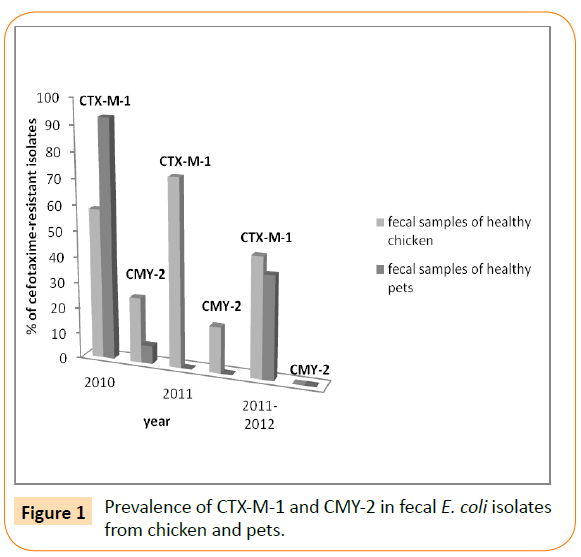
Figure 1: Prevalence of CTX-M-1 and CMY-2 in fecal E. coli isolates from chicken and pets.
In the same period (2012), Ben Sallem et al. have reported the isolation of cefotaxime-resistant E. coli isolates from 11 out of 80 fecal samples of healthy food-producing animals analysed (13.8%) [25]. Samples where chicken (17 samples), sheep (9 samples), cow (6 samples), horse (1 sample) dromedary (1 sample), and rabbit (3 samples). Cefotaxime-resistant isolates were from chicken (10 isolates) and dromedary (1 isolate). Nine of these isolates harboured the blaCTX-M-1 gene, and two of them harbored the blaTEM-135 or blaTEM-1b genes. The remaining two E. coli isolates contained the blaCMY-2 gene (encoding the beta-lactamase CMY-2). In addition, the same investigators found the blaCTX-M-1 gene in 92.8% of cefotaxime-resistant isolates in fecal E. coli from healthy pets (cats and dogs) for the first time in Tunisia and in Africa (Table 1) [26]. In the period between 2011 and 2012, the blaCTX-M-1 gene has been also reported in faecal samples from healthy chickens and pets (Figure 1) [27]. In this study, they reported for the first time in Tunisia of CTX-M-15 enzyme in fecal swabs from pet dogs with low percentage (6.6% of detected ESBL). Moreover this study reported the first data on CTX-M-9 type ESBL in healthy chickens, despite the occurrence of this β-lactamase exclusively in a clinical strain of Enterobacter cloacae isolated from a stool culture in intensive care unit of the Military Hospital of Tunis in 2005 for the first time in Tunisia [28]. These findings are in concordance with the study of Mora and co-workers performed in Spain in 2010 describing the zoonotic potential of CTX-M-9-producing avian isolates [29]. It seems that the expansion of CTX-M enzymes remains in evolution in Tunisian’s livestock. Indeed, more recently, in 2015, our team reported the occurrence of blaCTX-M-1 gene, one of them coharbored the TEM-1 enzyme, in 16 out of 17 cefotaxime-resistant E. coli isolates recovered from feces of healthy poultry from two avian husbandries [30].
It is noteworthy, that all Tunisian reports showed the high percentage of faecal carriage of ESBL-positive E. coli isolates from healthy food-producing animals and from pets. On the other hand, these Tunisian reports confirmed the prevalence of CTX-M-1 β-lactamase in food-producing animals and indicated that the poultry food (particularly chicken samples) and pets play an important role as a reservoir of cefotaxime-resistance genes. This finding is in agreement with that of previous studies carried out in France, Great Britain, Belgium, Portugal, Spain and China showing increased raw poultry meat colonized by CTX-M-1-producing E. coli isolates [13,31-36].
Despite the predominance of CTX-M-15 enzyme in human patients in Tunisia during the last decade, it seems that the emergence of CTX-M-1 in poultry isolates has influenced the epidemiology of CTX-M type enzymes in healthy humans in Tunisia (Table 2) [37-50]. This emergence might alarm for possible increase of blaCTX-M-1 over blaCTX-M-15 in Tunisian human isolates. Indeed, a recent study demonstrated the occurrence of blaCTX-M-1 genes-producing E. coli in 7.3% of Tunisian healthy humans [51]. The authors have warned for possible dissemination of such poultry isolates to human. This hypothesis is in concordance with previous studies performed in the Netherlands showing clonal relationships of blaCTXM- 1-producing E. coli isolates from poultry and humans [52]. The dissemination of such strains has likely happened through the food chain. However, it seems that this phenomenon of poultry to human dissemination might slightly influence the occurrence of blaCTX-M genes in human isolates. This statement is consistent with a recently completed survey of E. coli carrying ESBL in broiler chickens in Great Britain indicating the predominance of CTX-M-1, despite the widespread prevalence of E. coli CTX-M-15 producers among human populations [35,53].
| References |
Species concerned |
Detected ESBL enzymes (number of isolates, or frequencies, remakes) |
| Rhimi et al. 2002 |
6 Klebsiellapneumoniae
1 Proteusmirabilis
3 Salmonella Livingstone |
ACC-1 |
| Makanera et al. 2003 |
31 SalmonellaMbandaka |
TEM-4 (13 isolates); SHV-2a (16 isolates); ACC-1a (18 isolates) |
| Ben-Hamouda et al. 2004 |
49 K. pneumoniae |
SHV-12; SHV-2a |
| Bouallegue et al. 2005 |
16 Salmonella Livingstone |
CTX-M-27 (all isolates) |
| Ktari et al.2006 |
11 K. pneumoniae |
VIM-4; CTX-15; CMY-4 (Clonal isolates harbored the three genes) |
| Mamlouket al.2006 |
35 E. coli
27 K. pneumoniae |
CTX-M-15 (31 E. coli and 24 K. pneumoniae isolates, the majority of isolates of both species were clonal) ; CTX-M-16 (4 E. coli and 3 K. pneumoniae , clonal isolates) |
| Abbassi et al. 2008 |
9 K. pneumoniae
2 E. coli |
SHV-11 (1 K. pneumoniae ) ;SHV-27 (one K. pneumoniae); CTX-M-15 ( all isolates) |
| Ben Achour et al. 2009 |
1 K. pneumoniae |
TEM-164 |
| Ben Achour et al. 2009 |
1 K. pneumoniae |
CTX-M-28 |
| Bourouis et al. 2009 |
1Enterobacter cloacae |
CTX-M-9 |
| Ktari et al. 2009 |
112 SalmonellaLivingstone |
SHV-2a (2 isolates) ; ACC-1 (111 isolates; clonal isolates) |
| Ben Slama et al. 2011 |
14 E. coli |
CTX-M-15 (12 isolates); CTX-M-14a (1 isolate) ; CTX-M-14b (1 isolate) |
| Dahmen et al. 2010 |
45 K. pneumoniae
1 Citrobacterfreundii
3 Morganellamorgani
2 Providenciarettgeri
31 E. coli
16 E. cloacae
2 K. oxytoca |
CTX-M-15 (45 K. pneumoniae, 31 E. coli, 7 E. cloacae, 3 M. morgani) ; SHV-2a (2 E. cloacae) ; SHV-12 (9 E. cloacae, 2 K. oxytoca, 2 P. rettgeri) |
| Elhani et al. 2010 |
103 K. pneumoniae |
CTX-M-15 (43 isolates) ; CTX-M-14 (2 isolates) ; CTX-27 (2 isolates) ; SHV-12 (58 isolates); SHV-2a (3 isolates) |
| Ben Sallem et al. 2012 |
11 E. coli |
CTX-M-1 (11 isolates) ; TEM-52c (1 isolate) |
| Hammami et al. 2012 |
44 E. cloacae |
CTX-M-15 (39 isolates) ; SHV-12 (6 isolates); SHV-27 (1 isolate) |
| Hammami et al. 2013 |
15 E. coli |
CTX-M-15 (13 isolates, not clonal isolates by PFGE); SHV-12 (2 isolates) |
| Alibi et al. 2015 |
118 K. pneumoniae |
SHV (89%), CTX-M (81.35%) TEM (56.78%), (bla genes were not sequenced in all isolates) |
Table 2: The incidence of reported ESBL enzymes in Enterobacteriaceae isolates from patients and healthy human in Tunisia since (2000-2015).
Other ESBL-encoding genes have also been reported in E. coli of animal origin. Indeed, TEM-1, TEM-20 and TEM-135 β-lactamases were also present in association with CTX-Mtype ESBLs [12,17,25]. In the world, few studies reported the detection of TEM-20 ESBL type of animal origin [52].
The CMY type is the most frequently reported plasmidic AmpC beta-lactamases (pAmpC-BL) in E. coli [32]. In Tunisia, E. coli strains carrying the plasmid-borne blaCMY-2 gene have been sources of concern in animal production and public health. In fact, previous studies demonstrated high occurrence of CMY-2 AmpC-BL from different poultry farms, healthy pet animals and food-producing animals with percentages of 25.3%, 7.1% and 18% of cefotaximeresistant strains, respectively (Figure 1) [21,25,26]. In the last few years, different reports alerted about the dissemination of ESBL/ CMY-producing E. coli from food animals and healthy companion animals in different countries [9,13,32,34,54].
Clonality of ESBL-producing E. coli isolates and incompatibility groups of plasmid-borne ESBL-encoding genes
All cefotaxime-resistant E. coli isolates (including ESBL-and/or pAmpC-BL-producing strains) showed unrelated PFGE patterns, in the majority of Tunisian reports. This high genetic diversity detected indicated that the spread of blaCTX-M-1 and blaCMY-2 genes is not due to the dissemination of clonal strains but might reflect the spread of transferable plasmids. Indeed, Mnif et al. have reported the occurrence of blaCTX-M-1 and blaCMY-2 genes on IncI1 plasmids in the majority of studied strains, and also on IncK plasmids [21]. Moreover, Jouini et al. have detected the presence of IncI1 plasmids in blaCTX-M-1-containing isolates and IncK plasmids in blaCTX-M-14 and blaCTX-M-1-producing isolates [55]. These plasmids might carry the aforementioned genes, despite molecular confirmation has not been carried out. In the meantime, another study performed in Tunisia highlighted the dissemination of blaCTX-M-1 and blaCTX-M-9 genes located on IncI1 conjugative plasmids, whereas the blaCTX-M-15 gene was carried on IncFII plasmid [27]. More Recently in 2014, Ben Sallem et al. showed the detection of IncI1 and IncF replicon plasmids in combination with other replicons in CTX-M-1 and CMY-2- producing E. coli isolates obtained from healthy humans and animals [56]. These findings agreed with those of previous studies that showed the high prevalence of IncI1 plasmid carrying blaCTX-M and blaCMY-2 genes in ESBL producing- E. coli strains from animal origin [7,13,33,35,36,52].
Genetic environment of blaCTX-M genes
The investigation of genetic environments of blaCTX-M genes showed the presence of the ISEcp1-blaCTX-M-1-orf477 structure in the majority of E. coliisolates recovered both from food-producing animals and faecal samples of healthy pets [12,17,25,26]. Nevertheless, the ISEcp1 sequence was found truncated by the IS26 and IS5 elements, located in complementary orientation [17,27]. ISEcp1-blaCTX-M-14-IS903 structure was presented in two isolates recovered from chicken samples [17]. As regards the blaCMY-2 gene, the ISEcp1-blaCMY-2 structure was identified in faecal samples of healthy food-producing animals [25]. However, a novel sequence has been reported where the IS10 sequence was demonstrated in one isolate recovered from chicken sample, truncating the ISEcp1 sequence (Accession number JX440359). Several previous studies have reported the presence of the ISEcp1 element upstream of blaCTX-M-1 and blaCMY-2 genes in Enterobacteriaceae and indicated that ISEcp1 plays an important role in the capture, expression, and continuous mobilization of these genes [33,57,58].
All these reported findings are undoubtedly of great interest to the scientific community in Tunisia and in other parts of the world. However, the main shortcomings in the studies conducted are the small number of samples analysed, therefore, further studies based on representative sample sizes for each animal species are needed to determine the true prevalence and the precise epidemiology of ESBL production in Tunisia. It will be certainly of great interest to realize a large epidemiological study on ESBL production in industrial husbandry, especially poultry industry, in human population, in agriculture (irrigation water, soil, and vegetables) as well as in wastewater or aquatic environments in order to assess the genetic epidemiology of circulating ESBLproducer isolates and the mobile genetic vehicles of blaSHV genes.
Conclusion
In Tunisia, cephalosporin-resistance by production of ESBLs enzymes of CTX-M group and CMY-type are prevalent especially from poultry. Chicken husbandry was identified as the main food sector contributing to the ESBL with other antibiotic resistance genes reservoir in animals that may be acquired by humans through handling or consumption of contaminated meat. In addition, the dissemination of blaCTX-M gene may spread differently in the future owing to increasing plasmid exchanges between the two hosts. Taken together, this situation is therefore alarming and needs establishment of continuous monitoring program of antimicrobial resistance in Tunisia.
Acknowledgements
Authors would like to acknowledge Pr. Carmen Torres (Area de Bioquímica y Biología Molecular, Universidad de La Rioja, Logroño, Spain) for her continuous support to Tunisian researchers in the field of antimicrobial resistance and for her enormous efforts in the majority of published works investigating ESBL-producing bacteria in Tunisia.
Conflict of Interest
Mahrouki Sihem and Abbassi Mohamed Salah have contributed equally to the realization of this work. None of the contributing authors has any conflict of interests relevant to the subject matter or materials discussed in the manuscript. No funding or other financial support was received.
6499
References
- Sun Y, Zeng Z, Chen S, Ma J, He L, et al. (2010) High prevalence of bla(CTX-M) extended-spectrum β-lactamase genes in Escherichia coli isolates from pets and emergence of CTX-M-64 in China. ClinMicrobiol Infect 16: 1475-1481.
- Carattoli A (2008) Animal reservoirs for extended spectrum beta-lactamase producers. ClinMicrobiol Infect 14 Suppl 1: 117-123.
- Gupta K, Scholes D, Stamm WE (1999) Increasing prevalence of antimicrobial resistance among uropathogens causing acute uncomplicated cystitis in women. JAMA 281: 736-738.
- Briñas L, Moreno MA, Zarazaga M, Porrero C, Sáenz Y, et al. (2003) Detection of CMY-2, CTX-M-14, and SHV-12 beta-lactamases in Escherichia coli fecal-sample isolates from healthy chickens. Antimicrob Agents Chemother 47: 2056-2058.
- Sáenz Y, Briñas L, Domínguez E, Ruiz J, Zarazaga M, et al. (2004) Mechanisms of resistance in multiple-antibiotic-resistant Escherichia coli strains of human, animal, and food origins. Antimicrob Agents Chemother 48: 3996-4001.
- Smet A, Martel A, Persoons D, Dewulf J, Heyndrickx M, et al. (2008) Diversity of extended-spectrum beta-lactamases and class C beta-lactamases among cloacal Escherichia coli Isolates in Belgian broiler farms. Antimicrob Agents Chemother 52: 1238-1243.
- Blanc V, Mesa R, Saco M, Lavilla S, Prats G, et al. (2006) ESBL- and plasmidic class C beta-lactamase-producing E. coli strains isolated from poultry, pig and rabbit farms. Vet Microbiol 118: 299-304.
- Li XZ, Mehrotra M, Ghimire S, Adewoye L (2007) beta-Lactam resistance and beta-lactamases in bacteria of animal origin. Vet Microbiol 121: 197-214.
- Winokur PL, Vonstein DL, Hoffman LJ, Uhlenhopp EK, Doern GV (2001) Evidence for transfer of CMY-2 AmpC beta-lactamase plasmids between Escherichia coli and Salmonella isolates from food animals and humans. Antimicrob Agents Chemother 45: 2716-2722.
- Khan AA, Nawaz MS, Summage West C, Khan SA, Lin J (2005) Isolation and molecular characterization of fluoroquinolone-resistant Escherichia coli from poultry litter. PoultSci 84: 61-66.
- Ponce-Rivas E, Muñoz-Márquez ME, Khan AA (2012) Identification and molecular characterization of class 1 integrons in multiresistant Escherichia coli isolates from poultry litter. Appl Environ Microbiol 78: 5444-5447.
- Slama BK, Jouini A, SallemBR, Somalo, S, Sáenz Y, et al. (2010) Prevalence of broad-spectrum cephalosporin-resistant Escherichia coli isolates in food samples in Tunisia, and characterization of integrons and antimicrobial resistance mechanisms implicated. Inter J Food Microbiol137: 281-286.
- Smet A, Martel A, Persoons D, Dewulf J, Heyndrickx M, et al. (2010) Broad-spectrum β-lactamases among Enterobacteriaceae of animal origin: molecular aspects, mobility and impact on public health. FEMS Microbiol Rev 34: 295-316.
- Abbassi MS, Torres C,Achour W,Vinue V,Sáenz Y, et al. (2008) Genetic characterisation of CTX-M-15-producing Klebsiellapneumoniae and Escherichia coli strains isolated from stem cell transplant patients in Tunisia. Inter J Antimicrobial Agents 32: 308-314.
- Alibi S,Ferjani A,Boukadida J (2015) Molecular characterization of extended spectrum beta-lactamases produced by Klebsiellapneumoniae clinical strains from a Tunisian Hospital. Med Mal Infect 45:139-143.
- Chouchani C, Marrakchi R, El Salabi A (2011) Evolution of β-lactams resistance in Gram-negative bacteria in Tunisia. Crit Rev Microbiol 37: 167-177.
- Jouini A, Vinué L, Slama KB, Sáenz Y, Klibi N, et al. (2007) Characterization of CTX-M and SHV extended-spectrum beta-lactamases and associated resistance genes in Escherichia coli strains of food samples in Tunisia. J AntimicrobChemother 60: 1137-1141.
- Hammerum AM, Jakobsen L, Olsen SS, Agersø Y (2012) Characterization of CTX-M-14- and CTX-M-15-producing Escherichia coli of porcine origin. J AntimicrobChemother 67: 2047-2049.
- Toszeghy M, Phillips N, Reeves H, Wu G, Teale C, et al. (2012) Molecular and phenotypic characterisation of extended spectrum β-lactamase CTX-M Escherichia coli from farm animals in Great Britain. Res Vet Sci 93: 1142-1150.
- Hiki M,Usui M, Kojima A, Ozawa M, Ishii Y, et al. (2013) Diversity of plasmid replicons encoding the blaCMY-2 gene in broad-spectrum cephalosporin-resistant Escherichia coli from livestock animals in Japan. Food Borne Pathog10: 243-249.
- Mnif B, Ktari S, Rhimi FM, Hammami A1 (2012) Extensive dissemination of CTX-M-1- and CMY-2-producing Escherichia coli in poultry farms in Tunisia. LettApplMicrobiol 55: 407-413.
- Meunier D, Jouy E, Lazizzera C, Kobisch M, Madec JY (2006) CTX-M-1- and CTX-M-15-type beta-lactamases in clinical Escherichia coli isolates recovered from food-producing animals in France. Int J Antimicrob Agents 28: 402-407.
- Sennati S, Santella G, Di Conza J, Pallecchi L, Pino M, et al. (2012) Changing epidemiology of extended-spectrum β-lactamases in Argentina: emergence of CTX-M-15. Antimicrob Agents Chemother 56: 6003-6005.
- Grami R, Dahmen S, Mansour W, Mehri W, Haenni M, et al. (2014) blaCTX-M-15-carrying F2:A-:B- plasmid in Escherichia coli from cattle milk in Tunisia. Microb Drug Resist 20: 344-349.
- Ben Sallem R, Ben Slama K, Sáenz Y, Rojo-Bezares B, Estepa V, et al. (2012) Prevalence and characterization of extended-spectrum beta-lactamase (ESBL)- and CMY-2-producing Escherichia coli isolates from healthy food-producing animals in Tunisia. Foodborne Pathog Dis 9: 1137-1142.
- Sallem RB, Gharsa H, Slama KB, Rojo-Bezares B, Estepa V, et al. (2013) First detection of CTX-M-, CMY-2, and QnrB19 resistance mechanisms in fecal Escherichia coli isolates from healthy pets in Tunisia. Vector Borne Zoonotic Dis 13: 98-102.
- Grami R, Mansour W, Dahmen S, Mehri W, Haenni M, et al. (2013) The blaCTX-M-1 IncI1/ST3 plasmid is dominant in chickens and pets in Tunisia. J AntimicrobChemother 68: 2950-2952.
- Bourouis A, Dubois V, Coulange L, André C, Bejhadj C, et al. (2011) First report of CTX-M-9 in a clinical isolate of Enterobacter cloacae in a Tunisian hospital. PatholBiol (Paris) 59: 187-191.
- Mora A, Herrera A,Mamami R, Lopez C, Alonzo M.P, et al. (2010) Recent emergence of clonal group O25b:K1:H4-B2-ST131 ibeA strains among Escherichia coli poultry isolated, including CTX-M-9-producing strains and comparison with clinical human strains. Appl Environ Microbiol 76: 6991-6997.
- Kilani H,Abbassi MS,Ferjani S,Mansouri R,Sghaier S, et al. (2015) Occurrence of blaCTX-M-, qnrB1 and virulence genes in avian ESBL-producing Escherichia coli isolates from Tunisia. Front Cell Infect Microbiol 5: 38. doi: 10.3389/fcimb.2015.00038.
- Egea P, López-Cerero L, Torres E, Gómez-Sánchez Mdel C, Serrano L, et al. (2012) Increased raw poultry meat colonization by extended spectrum beta-lactamase-producing Escherichia coli in the south of Spain. Int J Food Microbiol 159: 69-73.
- Doi Y, Paterson DL, Egea P, Pascual A, López-Cerero L, et al. (2010) Extended-spectrum and CMY-type beta-lactamase-producing Escherichia coli in clinical samples and retail meat from Pittsburgh, USA and Seville, Spain. ClinMicrobiol Infect 16: 33-38.
- Girlich D, Poirel L, Carattoli A, Kempf I, Lartigue MF, et al. (2007) Extended-spectrum beta-lactamase CTX-M-1 in Escherichia coli isolates from healthy poultry in France. Appl Environ Microbiol 73: 4681-4685.
- Pitout JD, Laupland KB (2008) Extended-spectrum beta-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect Dis 8: 159-166.
- Randall LP, Clouting C, Horton RA, Coldham NG, Wu G, et al. (2011) Prevalence of Escherichia coli carrying extended-spectrum β-lactamases (CTX-M and TEM-52) from broiler chickens and turkeys in Great Britain between 2006 and 2009. J AntimicrobChemother 66: 86-95.
- Zheng H, Zeng Z, Chen S, Liu Y, Yao Q, et al. (2012) Prevalence and characterisation of CTX-M β-lactamases amongst Escherichia coli isolates from healthy food animals in China. Int J Antimicrob Agents 39: 305-310.
- Ben Achour N, Mercuri PS, Ben Moussa M, Galleni M, Belhadj O (2009) Characterization of a novel extended-spectrum TEM-type beta-lactamase, TEM-164, in a clinical strain of Klebsiellapneumoniae in Tunisia. Microb Drug Resist 15: 195-199.
- Ben Achour N, Mercuri PS, Power P, Belhadj C, Ben Moussa M, et al. (2009) First detection of CTX-M-28 in a Tunisian hospital from a cefotaxime-resistant Klebsiellapneumoniae strain. PatholBiol (Paris) 57: 343-348.
- Ben-Hamouda T, Foulon T, Ben-Mahrez K (2004) Involvement of SHV-12 and SHV-2a encoding plasmids in outbreaks of extended-spectrum beta-lactamase-producing Klebsiellapneumoniae in a Tunisian neonatal ward. Microb Drug Resist 10: 132-138.
- Ben Slama K, Ben Sallem R, Jouini A, Rachid S, Moussa L, et al. (2011) Diversity of genetic lineages among CTX-M-15 and CTX-M-14 producing Escherichia coli strains in a Tunisian hospital. CurrMicrobiol 62: 1794-1801.
- Bouallègue-Godet O, Ben Salem Y, Fabre L, Demartin M, Grimont PA, et al. (2005) Nosocomial outbreak caused by Salmonella enterica serotype Livingstone producing CTX-M-27 extended-spectrum beta-lactamase in a neonatal unit in Sousse, Tunisia. J ClinMicrobiol 43: 1037-1044.
- Dahmen S,Bettaieb D, Mansour W,Boujaafar N,Bouallègue O, et al. (2010) Characterization and molecular epidemiology of extended spectrum β-lactamases in clinical isolates of Enterobacteriaceae in a Tunisian University Hospital. Microb Drug Resist 16:163-170.
- Elhani D,Bakir L,Aouni M,Passet V,Arlet G, et al. (2010) Molecular epidemiology of extended-spectrum β-lactamase producing Klebsiella pneumonia strains in a university hospital in Tunis, Tunisia, 1999-2005. ClinMicrobiol Infect 16: 157-164.
- Hammami S, Saidani M, Ferjeni S, Aissa I, Slim A, et al. (2013) Characterization of extended spectrum β-lactamase-producing Escherichia coli in community-acquired urinary tract infections in Tunisia. Microb Drug Resist 19: 231-236.
- Hammami S, BoubakerBBI,Saidani M,Lakhal E, Hassen BA, et al. (2012) Characterization and molecular epidemiology of extended spectrum beta-lactamase producing Enterobacter cloacae isolated from a Tunisian hospital. Microb Drug Resist 18: 59-65.
- Ktari S,Arlet A,Verdet C,Jaoua S,Kachrid, A, et al. (2009) Molecular epidemiology and genetic environment of acquired blaACC-1 in Salmonella enteric serotype Livingstone causing a large nosocomial outbreak in Tunisia. Mirob Drug Resist 15: 279-286.
- Ktari S,Arlet G,Mnif B, Gautier V,MahjoubiF, et al. (2006) Emergence of multidrug-resistant Klebsiellapneumoniae isolates producing VIM-4 metallo-β-lactamase, CTX-M-15 extended-spectrum β-lactamase, and CMY-4 AmpC β-lactamase in a Tunisian University Hospital. Antimicrob Agents Chemother 50: 4198-4201.
- Makanera A,Arlet G, Gautier V,ManaiM (2003) Molecular epidemiology and characterization of plasmid-encoded β-lactamases produced by Tunisian clinical isolates of Salmonella enterica serotype Mbandaka resistant to broad-spectrum cephalosporins. J ClinMicrobiol 41: 2940-2945.
- Mamlouk K, Boutiba-Ben Boubaker I, Gautier V, Vimont S, Picard B, et al. (2006) Emergence and outbreaks of CTX-M beta-lactamase-producing Escherichia coli and Klebsiellapneumoniae strains in a Tunisian hospital. J ClinMicrobiol 44: 4049-4056.
- Rhimi-Mahjoubi F, Bernier M,Arlet G, Ben Jemaa Z,Jouve P, et al. (2002)Mise en evidence de la cephalosporinaseplasmidique ACC-1 dansdifferentesenterobacteries (Klebsiellapneumoniae, Proteus mirabilis, Salmonella) isoleesdans un hôpitalTunisien (Sfax 1997–2000). PatholBiol 50: 7-11.
- Ben Sallem R, Ben Slama K, Estepa V, Jouini A, Gharsa H, et al. (2012) Prevalence and characterisation of extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli isolates in healthy volunteers in Tunisia. Eur J ClinMicrobiol Infect Dis 31: 1511-1516.
- Leverstein-van Hall MA, Dierikx CM, Cohen Stuart J, Voets GM, van den Munckhof MP, et al. (2011) Dutch patients, retail chicken meat and poultry share the same ESBL genes, plasmids and strains. ClinMicrobiol Infect 17: 873-880.
- Canton R, Gonzales-Alba JM, Galan RC (2012) CTX-M-enzymes: Origin and diffusion. Frontiers. Microb 3: 110.
- Smet A, Martel A,Persoons D,Dewulf J,Hevndrickx M, et al. (2009)Cloeckaert, A, Comparative analysis of extended-spectrum-b-lactamase (ESBL)-carrying plasmids from different members of Enterobacteriaceae isolated from poultry, pigs and humans: evidence for a shared β-lactam resistance gene pool?. J AntimicrobChemoth63: 1286-1288.
- Jouini A, Slama KB, Klibi N, Sallem RB, Estepa V, et al. (2013) Lineages and virulence gene content among extended-spectrum β-lactamase-producing Escherichia coli strains of food origin in Tunisia. J Food Prot 76: 323-327.
- Ben Sallem R, Ben Slama K, Rojo-Bezares B, Porres-Osante N, Jouini A, et al. (2014) IncI1 plasmids carrying bla(CTX-M-1) or bla(CMY-2) genes in Escherichia coli from healthy humans and animals in Tunisia. Microb Drug Resist 20: 495-500.
- Lim SK, Lee HS, Nam HM, Jung SC, Bae YC (2009) CTX-M-type beta-lactamase in Escherichia coli isolated from sick animals in Korea. Microb Drug Resist 15: 139-142.
- Eckert C, Gautier V, Arlet G (2006) DNA sequence analysis of the genetic environment of various blaCTX-M genes. J AntimicrobChemother 57: 14-23.






