Key words
STEC, PCR, mutton, chicken, shiga toxin, mTSB and mEC.
Introduction
Strains of shiga-toxingenic E. coli (STEC) of different serotypes are recognized as important human pathogens of animal origin and can cause severe diseases such as Haemmorhagic Colitis, Haemolytic Uraemic Syndrome and Thrombotic Thrombocytopaenic Purpura [5]. E. coli O157:H7 is an important serotype of STEC which accounts for 70-80% of recognized clinical diseases [6]. It has been reported that >200 serotypes of E. coli including O157 and non-O157 STEC produce shiga toxins stx1 and stx2 [12] which are also known as verotoxins.
E. coli strains carrying stx2 gene along with enterohaemolysin (hlyA) gene are potentially dangerous to human health. Shiga toxins bind to receptors on the bowel mucosa and are elaborated and translocated into the cell interior and inactivate ribosomal RNA leading to the inhibition of protein synthesis in cells expressing glycolipid G3b (globotriaosylceramide) and eventually causes death of host cells [11,14,18]. High levels of G3b are found in human kidney which is 1000 times more sensitive to the cytotoxic action of stx2 than that of stx1 [10].
Blood released due to mucosal damage is lysed by enterohaemolysin liberating heme and haemoglobin which facilitates rapid multiplication of the organism [1,13]. As multiplication takes place, further toxin production occurs, causing greater damage, releasing increasing amounts of blood and resulting in growth stimulation [9].
Epidemiological studies have shown that food of animal origin is the main source of human infection [7]. The organisms can enter human food chain from a number of intestinal contents before, during or after slaughter. That is why food of animal origin must be regarded as potential vehicle of STEC.
The present investigation was carried out for the presence of STEC in mutton and chicken samples collected from in and around Hyderabad, Andhra Pradesh.
Materials and Methods
A total of 100 meat samples (50 samples each of mutton and chicken) and 100 meat surface swabs (50 each of mutton and chicken swabs) were collected from freshly dressed and washed animal carcasses at slaughter houses and markets in Hyderabad. Meat samples (10gm each) and swabs were enriched in 90 ml of modified E. coli (mEC) broth and modified Tryptic Soy broth (mTSB) both supplemented with novobiocin at 37oC for 18 hours. An E. coli O157:H7 strain, obtained from National Institute of Enteric diseases, Kolkata was used as known positive strain in PCR analysis. All the enriched samples were subjected to PCR analysis for the presence of STEC by using oligonucleotide primers against three virulence genes i.e. stx1, stx2 and hlyA (Table 1) with an amplification size of 614, 779 and 361bp respectively as shown in figure 1 & 2.
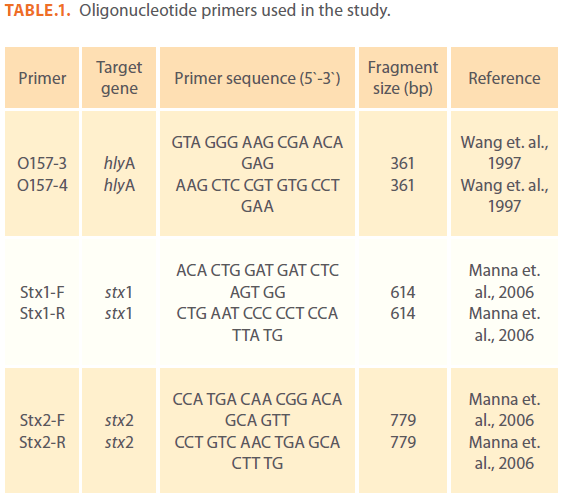
Table 1: Oligonucleotide primers used in the study.
1.5 ml of enriched broths were taken into eppendorf tubes and bacteria were were pelleted by centrifuging at 6000rpm for 5 min. and supernatant was discarded. To the pellet 50μl of molecular grade water was added and kept in a water bath at 65oC for 15 min. and snap chilled. Then centrifuge at 13000rpm for 5 min. and the supernatants were used as DNA templates for PCR analysis.
Bacterial DNA amplification was done in 20μl reaction mixture containing 2μl of 10X Taq DNA polymerase buffer (containing 100mM Tris with pH 9.0, 500mM KCl, 15mM MgCl2 and 1% Triton X-100), 2μl of 10mM of dNTP mix, 0.9U of Taq DNA polymerase (Genei), 2μl each of 4 p.moles/μl of forward and reverse primers and 5μl of crude bacterial cell lysate. Make this mixture to 20μl using molecular grade water. Amplification was done in thermal cycler following standardized conditions (Table 2).
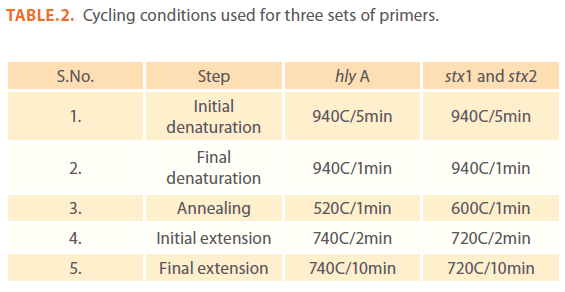
Table 2: Cycling conditions used for three sets of primers.
The amplified DNA fragments were resolved by agarose (1%) gel electrophoresis, stained with ethidium bromide (0.5μg/ml) and visualized with an UV transilluminator (Figure 1-4).
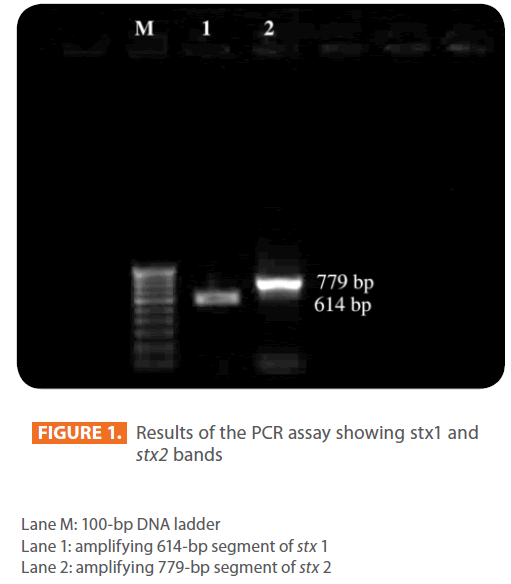
Figure 1: Results of the PCR assay showing stx1 and stx2 bands
Lane M: 100-bp DNA ladder
Lane 1: amplifying 614-bp segment of stx 1
Lane 2: amplifying 779-bp segment of stx 2
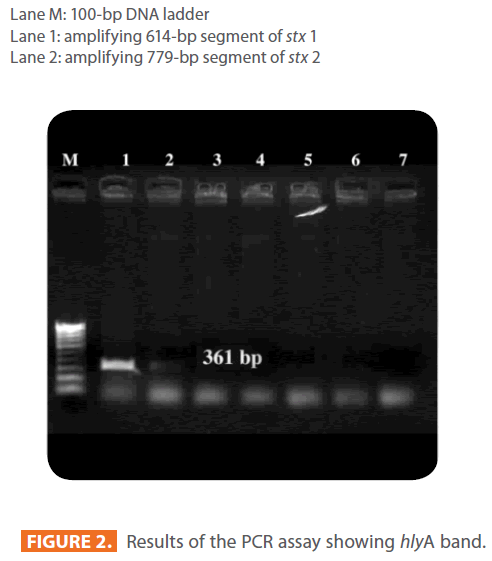
Figure 2: Results of the PCR assay showing hlyA band.
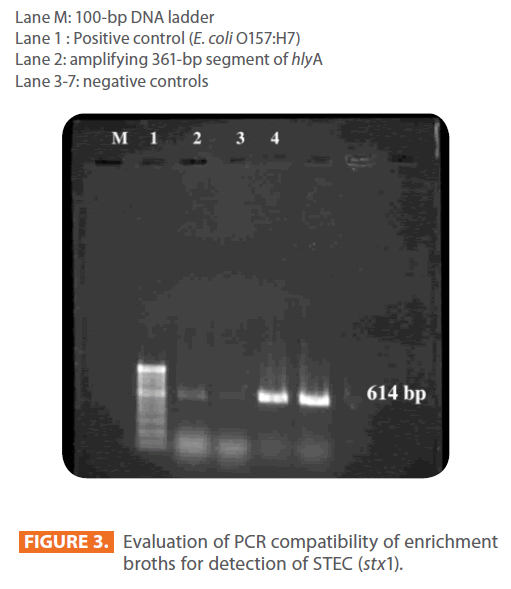
Lane M: 100-bp DNA ladder
Lane 1 : Positive control (E. coli O157:H7)
Lane 2: amplifying 361-bp segment of hlyA
Lane 3-7: negative controls
Figure 3: Evaluation of PCR compatibility of enrichment broths for detection of STEC (stx1).
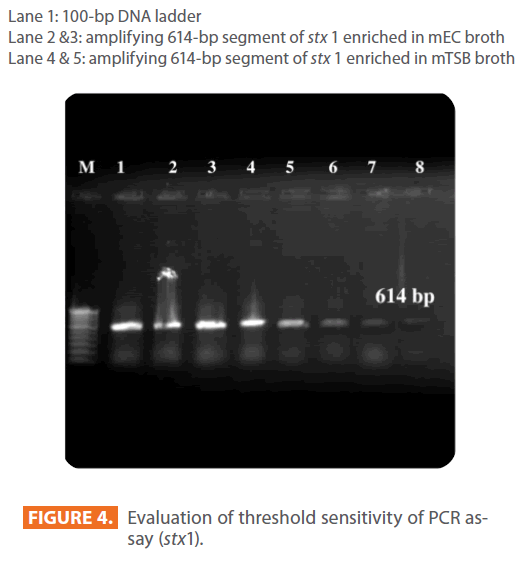
Lane 1: 100-bp DNA ladder
Lane 2 &3: amplifying 614-bp segment of stx 1 enriched in mEC broth
Lane 4 & 5: amplifying 614-bp segment of stx 1 enriched in mTSB broth
Figure 4: Evaluation of threshold sensitivity of PCR assay (stx1).
Sensitivity and Spiking studies: To know the sensitivity of PCR assay for detection of STEC, homogenized beef was inoculated with known E. coli O157:H7 strain in 10gm of mutton and transferred to two different enrichment media, mEC and mTSB broths. The PCR analysis was carried out after 10hr and 18hr of incubation period.
Results
The minimum detection level of STEC by PCR was 17cfu and 1.7cfu for mEC broth after 10hr and 18hr incubation respectively, where as it was 1.7cfu for mTSB after 10hr as well as 18hr incubation period. The results for sensitivity studies were depicted in Figure 4. The growth of STEC in samples as well as spiking studies was good in mTSB compared to mEC as shown in Figure 3.
The results for the presence of STEC in different meat samples are presented in Table 3. In this study, STEC were detected in 40% (20 out of 50) and 48% (24 out of 50) of mutton samples and mutton swabs respectively. Also STEC were detected in 14% (7 out of 50) of both chicken and chicken swab samples. Of the total 29% (58 out of 200) STEC positive samples (meat and meat swabs together), 72.4% (42 out of 58 STEC) samples showed presence of stx1, 27.5% of STEC (16 out of 58) showed stx2 and 13.7% of STEC (8 out of 58) showed presence of both stx1 and stx2.
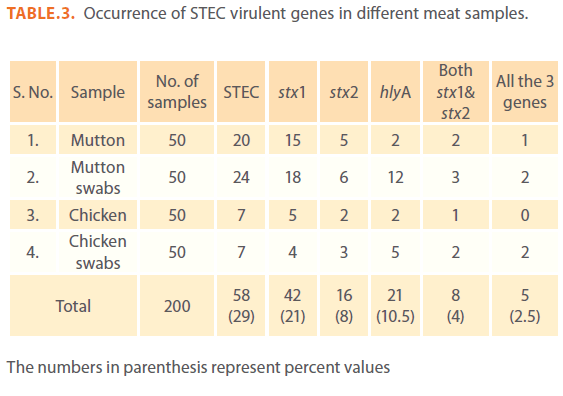
The numbers in parenthesis represent percent values
Table 3: Occurrence of STEC virulent genes in different meat samples.
When tested for presence of ‘hlyA’ by PCR, 36.2% (21 out of 58) of the total STEC positive samples showed presence of hlyA. The hlyA gene was detected in 10% (2 out of 20) of STEC positive mutton isolates, 50% (12 out of 24) mutton swab isolates, 28.5% (2 out of 7) chicken isolates and 71.4% (5 out of 7) chicken swab isolates. Among 58 STEC positive isolates, 8.6% (1 mutton, 2 mutton swab and 2 chicken swabs) isolates were possessing all the 3 virulent genes.
Discussion
STEC are the cause of acute infections in humans such as HC, HUS and thrombotic thrombocytopenic purpura [15]. They can also induce subclinical infections and mild diarrhea [7]. Foods of animal origin are the most common vehicle of STEC infection in humans. It has been isolated from wide variety of foods such as beef, mutton, chicken, pork, eggs, milk and milk products. Detection of STEC from food stuffs is problematic, since they are present at low level together with large number of competing microflora and also they may be injured by different food processing methods. Traditional methods for detecting and identifying STEC are labour intensive and time consuming. Hence there is a need to develop reliable and rapid methods for detection of STEC from foods.
Commercially available rapid antigen-antibody test methods such as ELISA and lateral flow immuno precipitation and molecular methods like PCR have reduced the time and labour involved in the analysis of food products. These assays still require enrichment of the food samples but significantly reduce the overall assay time compared with conventional methods [3]. In contrast to culture based methods, PCR methods may also detect cells that are non-culturable [4]. The polymerase chain reaction is a sensitive, rapid technique in which a few copies of target DNA can be amplified to a level detectable by gel electrophoresis [8]. The PCR represents a major advance in terms of the speed, sensitivity and specificity of diagnostic methods and has been increasingly used to identify several bacterial species from food and clinical samples [17].
In the present study, the sensitivity of PCR was found to be 1.7cfu after 18hr incubation period in both mEC and mTSB broths. The sensitivity and specificity of STEC isolates by PCR was better with mTSB compared to mEC broth. In this study, mutton samples yielded high percentage of STEC compared to chicken samples. Many studies have shown that sheep are major reservoir of STEC [16]. The stx1gene was found more frequently in this study compared to stx2. Most of the published test studies also revealed the higher production of stx1 than stx2 [2,16].
Since the E. coli strains carrying the shiga toxin genes along with enterohaemolysin (hlyA) gene are potentially dangerous to human health, PCR is a very specific and rapid method of detecting the virulent STEC. The detection of STEC in foods of animal origin in this study warrants that human diarrhoeal stools during foodborne outbreaks must be routinely examined for STEC.
Conclusion
This study emphasizes the prevalence of virulent STEC in mutton and chicken samples in and around Hyderabad, India. STEC are present in many raw foods at low levels and frequencies and keeping in view the very low infectious dose of E. coli O157 (< 10 viable bacteria), much greater efforts are required for detection of these STEC. The possible ways of entry of various E. coli serotypes could be handling of meat and meat products by adopting improper hygienic measures during handling and processing. Therefore, careful processing of meat and meat products is an important public health measure. Proper hygienic measures need to be adopted for handling various food products in order to bring down the entry of these organisms in to meat, thereby safeguarding the human beings from health hazards.
196
References
- Beutin L, Montenegro M.A, Orskov I, Orskov F, Prada J, Zimmermenn S, Stephen R. (1989) Close association of verotoxin (Shiga like toxin) production with enterohemolysin production in strains of Escherichia coli. J ClinMicrobiol 27:2559- 2564.
- Cerqueira A.M.F, Guth B.E.C, Joaquim R.M, Andrade J.R.C. (1999) High occurrence of shiga toxin-producing Escherichia coli (STEC) in healthy cattle in Rio de Janeiro State, Brazil. Veterinary Microbiology 70: 111-121.
- Feng P. (2001). Rapid methods for detecting foodborne pathogens. In: FDA Bacteriological Analytical Manual. Available online at https://www.cfsan.fda.gov/~ebam/bam-al.html. Accessed 10 December 2004.
- Fode-Vaughan K.A, Maki J.S, Benson J.A, Collins M.L.P. (2003) Direct PCR detection of Escherichia coli O157:H7. Letters in ApplMicrobiol 37: 239-243.
- Griffin P.M, Tauxe R.V. (1991) The epidemiology of infections caused by Escherichia coli O157, other enterohaemorrhagicE.coli, and the associated haemolyticuraemic syndrome. Epidemiol Rev 13: 60-98.
- Gyles C, Johnson R, Anli A, Ziebell K, Pierard D, Aleksic S, Boerlin P. (1998) Association of enterohaemorrhagicEscherichia coli haemolysin with serotypes of shiga like toxin producing Escherichia coli of human and bovine origins. Appl Environ Microbiol 64: 4134-4141.
- Karmali M.A. (1989) Infection by verocytotoxin-producing Escherichia coli. ClinMicrobiol Rev 2: 15-38.
- Lantz P.G, Hahnhagerdal B, Radstrom P. (1994) Sample preparation methods in PCR based detection of food pathogens. Trends in Food SciTechnol 5: 384-389.
- Law D, Kelly J. (1995). Use of heme and hemoglobin by Escherichia coli O157 and other shiga-like-toxin-producing E.coliserogroups. Infect Imm 63: 700-702.
- Louise C.B, Obrig T.G. (1995) Specific interaction of Escherichia coli O157:H7 derived shiga-like toxin II with human renal endothelial cells. J Infect Dis 172: 1397-1401.
- Manna S.K, Brahmane M.P, Das R, Manna Chandana and Batabyal S. (2006) Detection of Escherichia coli O157 in foods of animal origin by culture and multiplex polymerase chain reaction. J Food SciTechnol 43(1): 77-79.
- Mead P.S, Griffin P.M. (1998) Escherichia coli O157:H7. Lancet 352: 1207-1212.
- Meng J, Doyle M.P. (1998) Microbiology of shiga toxin producing Escherichia coli in foods. In: Escherichia coli O157:H7 and other shiga toxin producing E.colistrains. Kaper J.B, O’Brien A.D.(eds)., ADM, Washington DC, p: 92-108.
- Nataro J.P, Kaper J.B. (1998) DiarrheagenicEscherichia coli. ClinMicrobiol Rev11: 142-201.
- O’Brien A.D, Holmes R.K. (1987) Shiga and Shiga-like toxins. Microbiol Rev 51: 206-220.
- Robins-Browne R.M. (1994) Escherichia coli strains that cause diarrhea. Models of bacterial pathogenesis. In: Recent advances in microbiology. The Australian Society for Microbiology, Melbourne, Australia 292-335.
- Sidjabat-Tambunan H, Bensink J.C, Bettelheim K.A. (1998) Isolaition of verocytotoxins-producing Escherichia coli from mutton carcasses. Aus Vet J 76(5): 364-365.
- Stone G.G, Oberst R.D, Hays M.P, McVey S, Chengappa M.M. (1994) Detection of Salmonella serovars from clinical samples by enrichment broth cultivation PCR procedure. J ClinMicrobiol 32: 1742-1749.
- Waddell T, Head S, Petric M, Cohen A, Lingwood C.A. (1988) Globotriosylceramide is specially recognized by the Escherichia coli verotoxin 2. BiochemBiophys Res Commun 152: 674-679.
- Wang R.F, Pao W.W, Cerniglia C.E. (1997 A universal protocol for PCR detection of 13 species of foodborne pathogen in foods. Journal of ApplMicrobiol 83: 727-736.












