Keywords
Phylogenetic tree; Antigenic shift; DNA mutation
Background
Avian influenza A virus is playing a key role to the emergence of human influenza. Recently transmission of Avian Influenza virus from bird to human has increased in several Asian countries. Influenza A virus is a member of Orthomyoxoviridae family is avirulent but it can be virulent by the acquisition of some genetic features which includes multibasic cleavage sites or glycosylation sites in the hemagglutinin (HA) gene can infect a wide range of species includes poultry, humans, horses, swine, quail etc. [1]. The highly pathogenic avian influenza (HPAI, strain type H5N1) virus has emerged in southern china more than a decade ago [2]. Among all A/Goose/Guangdong/1/1996 is the precursor of H5N1 viruses which is established initially in southern China from 1996 to 1999 in domestic geese [3,4]. From the emergence of this virus it has caused endemic infections in poultry industry in many Southeast Asian countries [5,6]. In HPAI virus there is a high rate of nucleotide substitution which is done by RNA virus [7]. RNA viruses have the higher rate of capability for mutation so that they can cross the species boundaries and jump to the new host to emerge new species [8]. It is believed that crossing of species boundaries require both environmental and appropriate virus genetics factors to transmission of the virus between species [9]. Segment 4 hemagglutinin (HA) genes is recognized to be the most mutable portion responsible for the attachment to the cell surface which acts as primary target of the host immune response resulting frequent genetic drift [10]. HPAI virus has the ability to transmit through both bird and human host contact system [11]. Variants from unique HPAI viruses could cause infection and has the ability to replicate in humans. Human strains may arise from some Hong Kong avian H5N1 strains without prior adaptation in a mammalian intermediate host [12] Avian virus strains circulate locally within poultry and wild birds. This virus may be migrated through the migratory birds to the new geographic regions. It can be spread by the movement of poultry and poultry products [13].
Virology
Avian influenza A consists of two major glycoproteins which are Hemaglutinin (HA) and Neuraminidase (NA) [14]. HA glycoproteins are more prone to attach to the cell surface sialic acid receptors. There is a difference between host surface receptors on the target cell which is believed to be the possible restrictive factor of avian influenza. HA gene of avian cell binds to Sia2-3Galactosecontaining receptor which is different from human Sia2- 6Galactose containing receptor [15]. Before functioning as a virus it needs post translational cleavage by host proteases [16]. HA followed by NA are important antigenic determinant from which neutralizing antibodies are directed. There are several subtypes of HA and NA. 18 different HA subtypes (H1 to H18) and 11 different NA subtypes (N1 to N11) are found [17]. There is a membrane protein named M2 protein which regulates the internal PH level of the virus. This membrane protein is responsible for uncoating the virus during early stages of viral replication [18]. Amantadine and rimantadine block this function. NA catalyze the cleavage of glycosidic linkages to sialic acid on the surface of the viral particle and host cell thus preventing the aggregation and facilitating the release of progeny viruses from the infected cell. Antiviral drugs like Oseltamivir and zanamivir (NA inhibitors) inhibits this important function are the key to the antiviral treatment.
Transmission
Transmission pattern of avian influenza A from one bird to another is poorly understood because of its complexity, huge number of species among birds and environmental factors. Some experiments have been done to identify the transmission pattern and it shows poorer transmission from infected to susceptible animals [19-21]. Migration process can influence transmission of viruses. Migratory birds can carry pathogens from country to country thereby playing a role distributing influenza viruses.
Materials and Methods: Sequence and Data Source
Data used in this study are obtained using nucleotide BLAST search from publicly available database of National Centre for Biotechnology Information (NCBI).Multiple sequence alignments, editing, assembly of strains were performed in windows platform with the Geneious program version 7.1.3 (trial). Numbers at nodes in the tree indicate Neighbor-Joining bootstraps value generated from 1,000 replicates.
Results and Discussion
In 2005 we have selected total 184 strains of Hemagglutinin (HA) strain of H5N1 (Figure 1 and Table 1).
| Accession number |
Strain name |
| DQ992752 |
Ck/GY/2173/05 |
| HQ664946 |
Ck/KH/022LC2b/05 |
| DQ992726 |
Dk/GX/4016/05 |
| DQ320912 |
Dk/HN/1608/05 |
| DQ992787 |
Dk/HN/5106/05 |
| DQ366306 |
Dk/VNM/1/05 |
Table 1: List of diverse strains from our analysis of 2005 are given below.
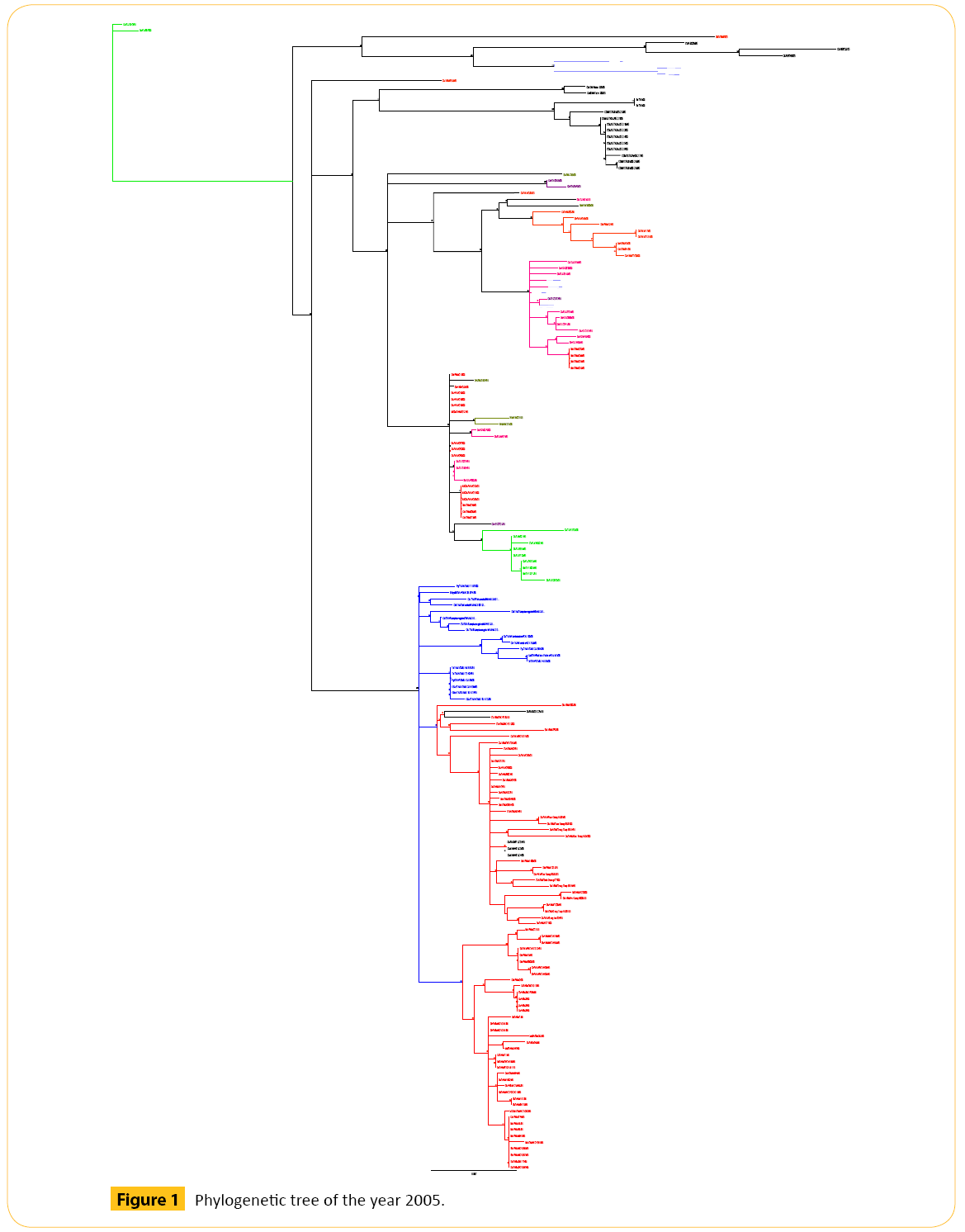
Figure 1: Phylogenetic tree of the year 2005.
In 2006 we have selected total 164 strains of Hemagglutinin (HA) strain of H5N1 (Figure 2 and Table 2).
| Accession number |
Strain name |
| HM172085 |
BHGs/QH/1-HVRI/06 |
| EF532626 |
Ck/IL/397/06 |
| DQ999872 |
Ck/THA/NP-172/06 |
| DQ992839 |
CMg/HK/645/06 |
| DQ992815 |
Gs/YN/1143/06 |
Table 2: List of diverse strains from our analysis of 2006 are given below.
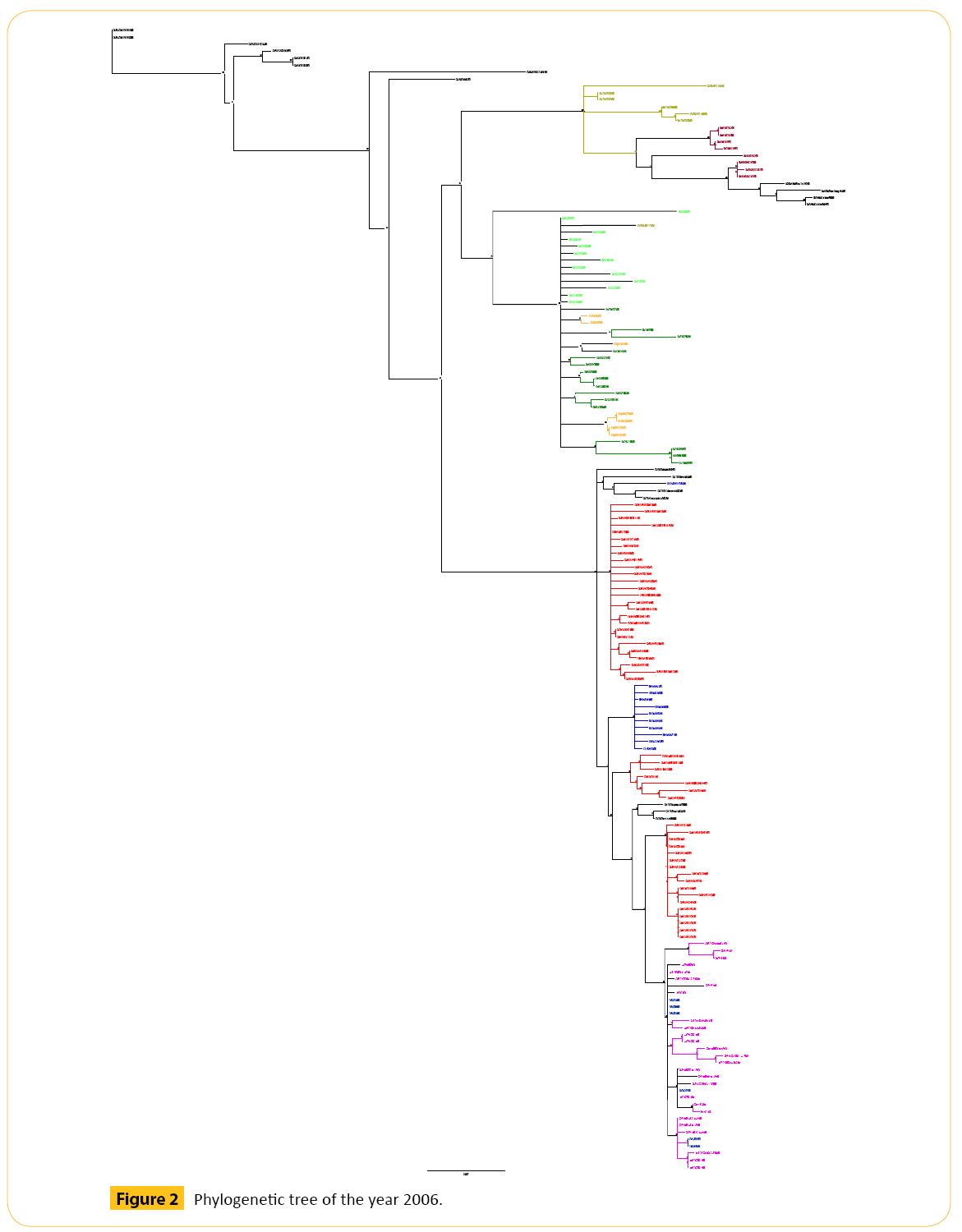
Figure 2: Phylogenetic tree of the year 2006.
In 2007 we have selected total 205 strains of Hemagglutinin (HA) strain of H5N1 (Figure 3 and Table 3).
| Accession number |
Strain name |
| CY091946 |
Dk/BT/BBVW-949-2D362/07 |
Table 3: List of diverse strains from our analysis of 2007 are given below.
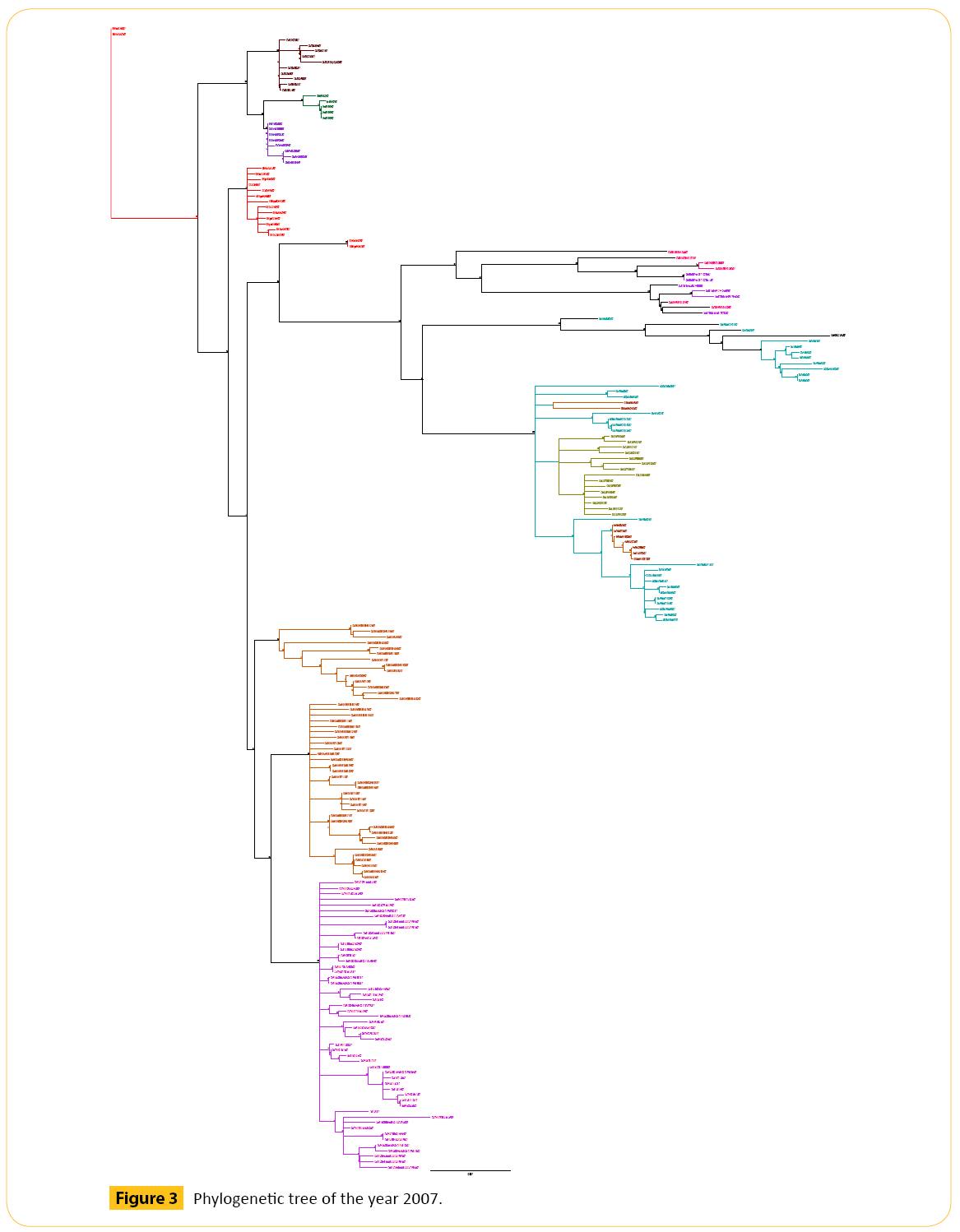
Figure 3: Phylogenetic tree of the year 2007.
From three years we have got some strains which seems to diverse from our analysis. We did our literature search but we did not get any information about these diverse strains. So it seems to us that these strains are not responsible for antigenic shift. Neighbor joining method and bootstrap value shows that these diverse strain is showing antigenic drift which may transfer to the other avian in the same country or other different country as well through migratory process.
From the phylogenetic tree of 2005 we came to see that all VNM like viruses formed a cluster. Among them Ck/KH/022LC2b/05 shows 81 bootstrap value with Ck/VNM/NCVD10/05. Number of bootstrap value is showing the similarities between these two strains. Strain Ck/KH/022LC2b/05 may be migrated or may be mutated which can show antigenic drift. Another Hunan like virus Dk/HN/5106/05 forms a clade with VNM like viruses. This strain can also show antigenic drift. One Guiyang (GY) virus Ck/ GY/2173/05 shows the genetic similarities with YN like viruses. Dk/GX/4016/05 and Dk/HN/1608/05 viruses are closely related to each other as they both are detached from their region and showing the same bootstrap value 57. One VNM virus (Dk/VNM/1/05) forming a clade with HB like viruses. Genetic character of this virus is different from other VNM like viruses as this is detached from the cluster of VNM like viruses showing the antigenic drift.
Phylogenetic tree analysis of 2006 shows that there are few diverse strains which may show genetic drift. One virus from Thailand (Ck/THA/NP-172/06) form clade with the GX like viruses. Two different viruses from two different area (CMg/HK/645/06 and Gs/YN/1143/06) are closely related to each other. They have the same bootstrap value. One QH like virus (BHGs/QH/1- HVRI/06) forming a clade with Turkey (TR) like viruses. Another Israel (IL) like virus (Ck/IL/397/06) forming a clade with Egypt (EG) like viruses. In 2006 there are almost 50 Nigeria (NGA) like viruses we have analyzed. But these viruses had not shown any genetic drift. They are forming different clades between them. Viruses from other countries like Egypt (EG), Qinghai (QH), Guangxi (GX), Thailand (THA), Combodia (KH) does not show any genetic drift.
In 2007 we did not see any genetic drift except one. One virus from Bantul (BT) (Dk/BT/BBVW-949-2D362/07) is closely related to the Badung (BDG) (Ck/BDG/BBVD-532/07) like virus. They show the same bootstrap value.
Our target is to identify the antigenic shift pattern from avian to human species. Analysis with amino acid (AA) shows the most specific way to identify the antigenic shift. For this we will combine the amino acid sequence of all these three years (2005, 2006, and 2007). After combining we had run alignment using ClustalW. It took almost 12 hours to complete.
The molecular mechanisms that enable avian influenza viruses to cross the species barrier and transmit efficiently in humans are incompletely understood. Some experiments have been done to identify the transmission pattern and it shows poorer transmission from infected to susceptible animals [22-24]. Migration process can influence transmission of viruses. Migratory birds can carry pathogens from country to country thereby playing a role distributing influenza viruses. Avian influenza A consists of two major glycoproteins which are Hemagglutinin (HA) and Neuraminidase (NA) [25]. HA glycoproteins are more prone to attach to the cell surface sialic acid receptors. There is a difference between host surface receptors on the target cell which is believed to be the possible restrictive factor of avian influenza.
Human infections are periodic. In some cases these viruses are accompanied by high mortality. As a result they are the major concern about the potential H5N1 as an endemic virus.
Although human infections are sporadic, they are accompanied by high mortality, raising major concerns about the potential of H5N1 as a pandemic virus [26]. Fortunately, H5N1 viruses have not yet naturally acquired the ability to stably transmit between humans [27,28]. One factor that limits transmission of avian viruses in humans is the receptor specificity of the hemagglutinin (HA) [29]. Avian viruses, like H5N1, preferentially bind to α2, 3 sialosides (avian-type receptors), whereas human viruses prefer α2, 6 sialosides (human-type receptors that are found in the human respiratory tract).
Before functioning as a virus it needs post translational cleavage by host proteases [30]. HA followed by NA are important antigenic determinant from which neutralizing antibodies are directed. There are several subtypes of HA and NA. 18 different HA subtypes (H1 to H18) and 11 different NA subtypes (N1 to N11) are found [31].
In humans, the SAα2, 6 Gal receptor is expressed mainly in the upper airway, while the SAα2, 3 Gal receptor is expressed in alveoli and the terminal bronchiole [32]. A virus with good affinity to both SAα 2, 3 Gal and SAα2, 6 Gal receptors may be a very dangerous one, which could both infect efficiently via its binding to Saα2, 6Gal in the upper airway and cause severe infection in the lung via its binding to Saα2, 3Gal.
Data used in this study are obtained inside using nucleotide BLAST search from publicly available database of National Centre for Biotechnology Information (NCBI). Multiple sequence alignments, editing, assembly of strains were performed in windows platform with the Geneious program version 7.1.3 (trial).
In this study we will analyze some avian hemagglutinin (H5N1) of different years. Analysis includes building nucleotide sequence and translating them into amino acid sequence. Then we will study amino acid positions with respect to some reported mutation to see the genetic pattern. After analyzing we will try to find out whether there are any similarities between avian and human or not. There are some reported avian H5N1 strains that affect human which are A/Goose/Hong Kong/739.2/2002 [33], A/ duck/Egypt/D1Br12/2007 [34], A/Duck/Singapore/3/97 [35], A/ egret/Egypt/1162/2006 [36]. All of these strain show preferential binding to Siaα (2, 6) Gal receptor that can infect a human. Few specific positions of amino acids are responsible for this binding.
We found some avian amino acid position Q222L [35], G224S (35), S227N [33], Q192H [34] are specific to SAα 2, 3 Gal receptor which has a previous reported history to affect human. On the other hand there are some avian amino acid position S227N [33,37], Q192H [34], N186K [37], Q196R [36], N182K [38], Q192R [38], S223N [39], G228S [36,40] are specific to SAα2, 6 Gal receptor which has a previous reported history to affect human. In our avian H5N1 analysis we did not find the exact location where reported mutations are occurred. But we found similar amino acid near the reported mutated position. We have analyzed around (before and after the mutation point) twenty positions with respect to the reported mutation point.
Here are the summary of some reported position which can be responsible for antigenic shift from avian to human (Tables 4 and 5).
| Amino acid mutation position with reference |
Short form |
Amino acid in Geneious in reported position |
| Ser-227-Asn [33,37] |
S227N |
P |
| Gln-192-His [34] |
Q192H |
W |
| Asn-186-Lys [37] |
N186K |
E |
| Gln-196-Arg [36] |
Q196R |
H |
| Asn-182-Lys [38] |
N182K |
N |
| Gln-192-Arg [38] |
Q192R |
W |
| Ser-223-Asn [39] |
S223N |
Q |
| Gly-228-Ser [40,36] |
G228S |
K |
| Gln-226-Leu [36] |
Q226L |
V |
Table 4: Reported a 2, 6 receptor specific avian amino acid position with mutation that can affect human.
| Amino acid mutation position with reference |
Short form |
Amino acid in Geneious in reported position |
| Gln-222-Leu [35] |
Q222L |
N |
| Gly-224-Ser [35] |
G224S |
R |
| Ser-227-Asn [33,37] |
S227N |
P |
| Gln-192-His [34] |
Q192H |
W |
Table 5: Reported a 2, 3 Receptor specific avian amino acid position with mutation that can affect human.
Analysis of our study data with respect to reported mutation point to see the antigenic shift pattern of avian H5N1 (Table 6).
| Reported AA with mutation point |
AA in Geneious in
reported position |
Exact AA which matches with the reported data with respect to specific mutation point (before 20 positions) |
Exact AA which matches with the reported data with respect to specific mutation point (after 20 positions) |
| S227N |
P227R (2)
P227A (1) |
S215
S219 |
S233P (3)
Polar to Nonpolar
S239 |
| Q192H |
W192 |
Q185R (3)
Q185H (2)
Q185K (2)
Polar to + electrically Charged (K, H, R) |
Q203
Q208 |
| N186K |
E186 |
N170G (5)
Polar to Nonpolar
N181T (2)
N182
N184 |
N184
N198K (2)
Polar to + electrically Charged (K)
N198S (1) |
| N182K |
N182 |
N162
N170G (5)
N171G (1)
Polar to Nonpolar (G)
N181T (2) |
N184D (1)
Polar to – electrically Charged (D)
N184S (1)
N198K (2)
Polar to + electrically Charged (K)
N198S (1) |
| Q192R |
W192 |
Q185K (2)
Q185R (3)
Q185H (2)
Polar to + electrically Charged (K, R, H) |
Q203
Q208 |
| S223N |
Q223 |
Q215
Q219 |
S233P (3)
Polar to Nonpolar (P)
S239 |
| G228S |
K228E (1)
K228N (1)
K228? (1) |
G217 |
G237
G240 |
Q226L |
V226E (1)
V226A (1) |
Q208 |
Q238 |
| Q196R |
H196 |
Q185K (2)
Q185R (3)
Q185H (2)
Polar to + electrically
Charged (K, R, H) |
Q203
Q208 |
Table 6: Correlation of Reported a 2, 6 receptor specific avian amino acid position with our experimental strain, and their mutation pattern (exact and around twenty positions): Original amino acid analysis.
In case of S227N (Ser-227-Asn) reported position, we found amino acid Proline (P) in our software (Geneious). We found two mutations here, Proline (P) to Arginine (R) in two strains and Proline (P) to Alanine (A) in one strain. We found amino acid Serine (S) in two positions (215 and 219) which are located within twenty positions before 227. In these two positions there are no mutations. We have also found that, there is Serine (S) in two positions (233 and 239) which are located within twenty positions after 227. We found S233P mutation in three strains which indicates that polarity is changed from polar to non-polar as Serine (S) is Polar and Proline (P) is nonpolar.
In case of Q192H (Gln-192-His) reported position, we found amino acid Tryptophan (W) in our software (Geneious). We found amino acid Glutamine (Q) in one position (185) which is located within twenty positions before 192. We found Q185R mutation in three strains, Q185H in two strains and Q185K in two strains. Here polarity is changed from Polar to positive electrically charged amino acid as Lysine (K), Histidine (H), Arginine (R) is positive electrically charged. We have also found that, there is Glutamine (Q) in two positions (203 and 208) which are located within twenty positions after 192. We found no mutation here.
In case of N186K (Asn-186-Lys) reported position, we found Glutamic acid (E) in our software (Geneious). We found Glutamic acid (E) in five positions (170, 171, 181, 182 and 184) which are located within twenty positions before 186. We found N170D mutation in numerous strains, N171S/D in numerous strains, N181T in two strains. We will not give importance to mutation in numerous mutations in one position as these are common and are not responsible for virulence. Here Polarity is not changed in case of N181T. We have also found that, there is Glutamic acid (E) in two positions (184 and 198) which are located within twenty positions after 186. We found N198K mutation in two strains and N198S in one strain which indicates that polarity is changed from polar to positively electrically charged Lysine (K) in case of N198K.
In case of N182K (Asn-182-Lys) reported position, we found Asparagine (N) in our software (Geneious). We found Asparagine (N) in four positions (162, 170, 171 and 181) which are located within twenty positions before 182. We found N170G in five strains, N171G in one strain and N181T in two strains. Here polarity is changed from Polar to non-polar amino acid as Glycine (G) is non-polar. We have also found that, there is Asparagine (N) in two positions (184 and 198) which are located within twenty positions after 182. We found N184D mutation in one strain, N184S mutation in one strain, N198S in one strain and N198K in two strains which indicates that polarity is changed from polar to negative electrically charged Aspartic acid (D), positive electrically charged Lysine (K) in case of N184D and N198K respectively.
In case of Q192R (Gln-192-Arg) reported position, we found Tryptophan (W) in our software (Geneious). We found Tryptophan (W) in one position (185) which is located within twenty positions before 192. We found Q185K in two strains, Q185R in three strains and Q185H in two strains. Here polarity is changed from Polar to positive electrically charged amino acid as Lysine (K), Histidine (H), Arginine (R) is positive electrically charged. We have also found that, there is Tryptophan (W) in two positions (203 and 208) which are located within twenty positions after 192.We found no mutation here.
In case of S223N (Ser-223-Asn) reported position, we found Glutamine (Q) in our software (Geneious). We found Glutamine (Q) in two positions (215 and 219) which is located within twenty positions before 223. We found no mutations here. We have also found that, there is Glutamine (Q) in two positions (233 and 239) which are located within twenty positions after 223. We found S233P mutation in three strains which indicate that polarity is changed from polar to non-polar as Proline (P) non-polar.
In case of G228S (Gly-228-Ser) reported position, we found amino acid Lysine (K) in our software (Geneious). We found two mutations here, Lysine (K) to Glutamic acid (E) in one strain and Lysine (K) to Asparagine (N) in one strain. We found Glycine (G) in one position (217) which is located within twenty positions before 228. We found no mutations here. We also found Glycine (G) in two positions (237 and 240) which is located within twenty positions before 228. We also found no mutations here.
In case of Q226L (Gln-226-Leu) reported position, we found amino acid Valine (V) in our software (Geneious). We found two mutations here, Valine (V) to Glutamic acid (E) in one strain and Valine (V) to Alanine (A) in one strain. We found Valine (V) in one position (208) which is located within twenty positions before 226. We found no mutations here. We also found Valine (V) in one position (238) which is located within twenty positions before 226. We also found no mutations here.
In case of Q196R (Gln-196-Arg) reported position, we found amino acid Histidine (H) in our software (Geneious). We found amino acid Glutamine (Q) in one position (185) which is located within twenty positions before 196. We found Q185R mutation in three strains, Q185H in two strains and Q185K in two strains. Here polarity is changed from Polar to positive electrically charged amino acid as Lysine (K), Histidine (H), Arginine (R) is positive electrically charged. We have also found that, there is Glutamine (Q) in two positions (203 and 208) which are located within twenty positions after 196. We found no mutation here.
In case of S227N (Ser-227-Asn) reported position, we found amino acid Proline (P) in our software (Geneious) (Table 7). We found two mutations here, Proline (P) to Arginine (R) in two strains and Proline (P) to Alanine (A) in one strain. We found amino acid Asparagine (N) in two positions (209 and 222) which are located within twenty positions before 227. We found N209R mutation in one strain and N222D in one strain. Here polarity is changed from Polar to positive electrically charged amino acid Arginine (R) and negative electrically charged Aspartic acid (D). We have also found that, there is Asparagine (N) in one position (236) which is located within twenty positions after 227. We found no mutation here.
| Reported AA with mutation point |
AA in Geneious in reported position |
Exact AA which matches with the reported data with respect to specific mutation point (before 20 positions) |
Exact AA which matches with the reported data with respect to specific mutation point (after 20 positions) |
| S227N |
P227R (2)
P227A (1) |
N209R (1)
Polar to + electrically
Charged (R)
N222D (1)
Polar to - electrically
Charged (D) |
S236 |
| Q192H |
W192 |
|
Q195
Q196 |
| N186K |
E186 |
K168N (2)
+ electrically Charged to polar (N)
K169R (1) |
|
| N182K |
N182 |
K168N (2)
+ electrically Charged to polar (N)
K169R (1) |
|
| Q192R |
W192 |
R178V (3)
+ electrically Charged to Nonpolar (V) |
R205G (1)
+ electrically Charged to Nonpolar (G) |
| S223N |
Q223 |
N209R (1)
Polar to + electrically Charged (R)
N222D (1)
Polar to - electrically Charged (D) |
S236 |
| G228S |
K228E (1)
K228N (1) |
G215
G219 |
S233P (3)
Polar to Nonpolar (P)
G239 |
| Q226L |
V226E (1)
V226A (1) |
L206I (4)
Q221
L225S (2)
Nonpolar to Polar (S)
L225F (1)
L225M (1) |
|
| Q196R |
H196 |
R178V (3)
+ electrically Charged to Nonpolar (V) |
R205G (1)
+ electrically Charged to Nonpolar (G) |
Table 7: Correlation of Reported a 2, 6 receptor specific avian amino acid position with our experimental strain, and their mutation
pattern (exact and around twenty positions): Mutated amino acid analysis.
In case of Q192H (Gln-192-His) reported position, we found amino acid Tryptophan (W) in our software (Geneious). We found no Histidine (H) which is located within twenty positions before 192. But we found Histidine (H) in two positions (195 and 196) which are located within twenty positions after 192. We found no mutation here.
In case of N186K (Asn-186-Lys) reported position, we found Glutamic acid (E) in our software (Geneious). We found amino acid Lysine (K) in two positions (168, 169) which are located within twenty positions before 186. We found K168N mutation in two strains, K169R in one strain. Here Polarity is changed from positive electrically charged to polar Asparagine (N) in case of K168N. We did not find any Lysine (K) which is located within twenty positions after 186.
In case of N182K (Asn-182-Lys) reported position, we found Asparagine (N) in our software (Geneious). We found Asparagine (N) in two positions (168, 169) which are located within twenty positions before 182. We found K168N in two strains and K169R in one strain. Here polarity is changed from positive electrically charged to polar Asparagine (N) in case of K168N. We did not found any Asparagine (N) which is located within twenty positions after 182.
In case of Q192R (Gln-192-Arg) reported position, we found Tryptophan (W) in our software (Geneious). We found Arginine (R) in one position (178) which is located within twenty positions before 192. We found R178V in three strains. Here polarity is changed from positive electrically charged to non-polar amino acid Valine (V). We have also found that, there is Arginine (R) in one position (205) which is located within twenty positions after 192. We found R205G in one strain. Here polarity is changed from positive electrically charged to non-polar amino acid Glycine (G).
In case of S223N (Ser-223-Asn) reported position, we found Glutamine (Q) in our software (Geneious). We found Asparagine (N) in two positions (209 and 222) which is located within twenty positions before 223. We found N209R in one strain and N222D in one strain. Here polarity is changed from polar to both positively charged amino acid Arginine (R) and negatively charged Aspartic acid (D). We have also found that, there is Asparagine (N) in one position (236) which is located within twenty positions after 223. We found no mutation here.
In case of G228S (Gly-228-Ser) reported position, we found amino acid Lysine (K) in our software (Geneious). We found two mutations here, Lysine (K) to Glutamic acid (E) in one strain and Lysine (K) to Asparagine (N) in one strain. We found Serine (S) in two positions (215 and 219) which is located within twenty positions before 228. We found no mutations here. We have also found Serine (S) in two positions (233 and 239) which is located within twenty positions before 228. We found S233P in three strains. Here polarity is changed from polar to non-polar amino acid Proline (P).
In case of Q226L (Gln-226-Leu) reported position, we found amino acid Valine (V) in our software (Geneious). We found two mutations here, Valine (V) to Glutamic acid (E) in one strain and Valine (V) to Alanine (A) in one strain. We found Leucine (L) in three positions (206, 221 and 225) which is located within twenty positions before 226. We found L206I in four strains, L225S in two strains, L225F in one strain, L225M in one strain. Here polarity is changed from non-polar to polar Serine (S) in case of L225S. We found no Leucine (L) which is located within twenty positions before 226.
In case of Q196R (Gln-196-Arg) reported position, we found amino acid Histidine (H) in our software (Geneious). We found amino acid Arginine (R) in one position (178) which is located within twenty positions before 196. We found R178V mutation in three strains. Here polarity is changed from positive electrically charged amino acid to non-polar amino acid Valine (V). We have also found that, there is Arginine (R) in one position (205) which is located within twenty positions after 196. We found R205G in one strain. Here polarity is changed from positive electrically charged amino acid to non-polar amino acid Glycine (G).
In case of Q222L (Gln-222-Leu) reported position, we found amino acid Asparagine (N) in our software (Geneious) (Table 8). We found one mutation here, Asparagine (N) to Aspartic acid (D) in one strain. We found amino acid Glutamine (Q) in two positions (203 and 208) which is located within twenty positions before 222. We found no mutation here. We have also found Glutamine (Q) in two positions (223 and 238) which is located within twenty positions after 222. We found no mutation here.
| Reported AA with mutation point |
AA in Geneious in reported position |
Exact AA which matches with the reported data with respect to specific mutation point (before 20 positions) |
Exact AA which matches with the reported data with respect to specific mutation point (after 20 positions) |
| Q222L |
N222D (1) |
Q203
Q208 |
Q223
Q238 |
| G224S |
R24K (2) |
G217 |
G237
G240 |
| S227N |
P227R (2)
P227A (1) |
S215
S219 |
S233P (3)
Polar to Nonpolar (P)
S239 |
| Q192H |
W192 |
Q185K (2)
Q185R (3)
Q185H (2)
Polar to + electrically Charged (K, R, H) |
Q203
Q208 |
Table 8: Correlation of Reportedα 2, 3 receptor specific avian amino acid position with our experimental strain, and their mutation pattern (exact and around twenty positions): Original amino acid analysis.
In case of G224S (Gly-224-Ser) reported position, we found amino acid Arginine (R) in our software (Geneious). We found one mutation here, Arginine (R) to Lysine (K) in two strains. We found amino acid Glycine (G) in one position (217) which is located within twenty positions before 224. We found no mutation here. We have also found Glycine (G) in two positions (237 and 240) which is located within twenty positions after 224. We found no mutation here.
In case of S227N (Ser-227-Asn) reported position, we found amino acid Proline (P) in our software (Geneious). We found two mutations here, Proline (P) to Arginine (R) in two strains and Proline (P) to Alanine (A) in one strain. We found amino acid Serine (S) in two positions (215 and 219) which are located within twenty positions before 227. In these two positions there are no mutations. We have also found that, there is Serine (S) in two positions (233 and 239) which are located within twenty positions after 227. We found S233P mutation in three strains which indicates that polarity is changed from polar to non-polar as Serine (S) is Polar and Proline (P) is non-polar.
In case of Q192H (Gln-192-His) reported position, we found amino acid Tryptophan (W) in our software (Geneious). We found amino acid Glutamine (Q) in one position (185) which is located within twenty positions before 192. We found Q185R mutation in three strains, Q185H in two strains and Q185K in two strains. Here polarity is changed from Polar to positive electrically charged amino acid as Lysine (K), Histidine (H), Arginine (R) is positive electrically charged. We have also found that, there is Glutamine (Q) in two positions (203 and 208) which are located within twenty positions after 192. We found no mutation here.
In case of Q222L (Gln-222-Leu) reported position, we found amino acid Asparagine (N) in our software (Geneious) (Table 9). We found one mutation here, Asparagine (N) to Aspartic acid (D) in one strain. We found amino acid Leucine (L) in two positions (206 and 221) which is located within twenty positions before 222. We found L206I in four strains. Here polarity is not changed. We have also found Leucine (L) in one position (225) which is located within twenty positions after 222. We found L225M in one strain, L225F in one strain and L225S in two strains. Here polarity is changed from non-polar to polar Serine (S) in case of L225S.
| Reported AA with mutation point |
AA in Geneious in reported position |
Exact AA which matches with the reported data with respect to specific mutation point (before 20 positions) |
Exact AA which matches with the reported data with respect to specific mutation point (after 20 positions) |
| Q222L |
N222D (1) |
L206I (4)
Nonpolar to Nonpolar (I)
Q221 |
L225M (1)
L225F (1)
L225S (2)
Nonpolar to Polar (S) |
| G224S |
R224K (2) |
G215
G219 |
S233P (3)
Polar to Nonpolar (P)
G239 |
| S227N |
P227R (2)
P227A (1) |
N209R (1)
Polar to + electrically Charged (R)
N222D (1)
Polar to - electrically Charged (D) |
S236 |
| Q192H |
W192 |
|
Q195
Q196 |
Table 9 :Correlation of Reported a 2, 3 receptor specific avian amino acid position with our experimental strain, and their mutation pattern (exact and around twenty positions): Mutated Amino Acid analysis.
In case of G224S (Gly-224-Ser) reported position, we found amino acid Arginine (R) in our software (Geneious). We found one mutation here, Arginine (R) to Lysine (K) in two strains. We found amino acid Serine (S) in two positions (215 and 219) which is located within twenty positions before 224. We found no mutation here. We have also found Serine (S) in two positions (233 and 239) which is located within twenty positions after 224. We found S233P in three strains. Here polarity is changed from polar to non-polar Proline (P).
In case of S227N (Ser-227-Asn) reported position, we found amino acid Proline (P) in our software (Geneious). We found two mutations here, Proline (P) to Arginine (R) in two strains and Proline (P) to Alanine (A) in one strain. We found amino acid Asparagine (N) in two positions (209 and 222) which are located within twenty positions before 227. We found N209R mutation in one strain and N222D in one strain. Here polarity is changed from Polar to positive electrically charged amino acid Arginine (R) and negative electrically charged Aspartic acid (D). We have also found that, there is Asparagine (N) in one position (236) which is located within twenty positions after 227. We found no mutation here.
In case of Q192H (Gln-192-His) reported position, we found amino acid Tryptophan (W) in our software (Geneious). We found no Histidine (H) which is located within twenty positions before 192. But we found Histidine (H) in two positions (195 and 196) which are located within twenty positions after 192. We found no mutation here.
Conflict of interest
None
5825
References
- Alipio BC, David FC, Isabel RS, Juan CNM. Manuel AE. Comparison of batch, stirred flow chamber, and column experiments to study adsorption, Desorption and transport of carbofuran within two acidic soils. Chemosphere. 2012; 88(1): 106-120.
- Arnaud B, Richard C, Michel S. A comparison of five pesticides adsorption and Desorption processes in thirteen contrasting field soils. Chemosphere. 2005; 61(5): 668-676.
- Chunxian W, Jin-Jun W, Su-Zhi Z, Zhong-Ming Z. Adsorption and Desorption of Methiopyrsulfuron in Soils. Pedosphere. 2011; 21(3): 380-388.
- Chunxian W, Suzhi Z, Guo N, Zhongming Z, Jinjun W. Adsorption and Desorption of herbicide monosulfuron-ester in Chinese soils. J Environ Sci. 2011; 23(9): 1524-1532.
- Christine MFB, Josette MF. Adsorption-desorption and leaching of phenylurea herbicides on soils. Talanta. 1996; 43(10): 1793-1802.








