Keywords
Phylogenetic tree; Sequence similarities; Antiviral resistance; DNA mutation
Abbreviations
SP: Singapore; SN: Swine; TN: Taiwan, HS: Helsinki
Introduction
The sudden emergence and spread of the swine-derived influenza virus from Mexico led to the displacement of the oseltamivirresistant seasonal H1N1 virus by the new A (H1N1) pdm 09 virus. However, given the awareness of oseltamivir resistance, much closer monitoring has been carried out, by both phenotypic testing by enzyme assay and sequencing. Viruses with the H274Y mutation have been detected in patients after treatment or prophylaxis with oseltamivir or peramivirand also in untreated patients. Resistance has been detected as early as 48 hours posttreatment [1-3].
Neuraminidase, also referred to as sialidase, is an antigenic glycoprotein anchored in the surface envelope of the influenza virions, which hydrolytically cleaves the terminal sialic acid from the host cell receptors. Thus, it plays a crucial role in the release of viral progeny from the membranes of infected cells, prevents self-aggregation of virions and facilitates the movement of the infectious viral particles in the mucus of the respiratory epithelia [4,5]
Influenza neuraminidase has been established as a key drug target for the prophylaxis and treatment of influenza infections, predominantly for the following reasons: Firstly, the structure of the influenza neuraminidase active site is highly conserved between influenza A and B strains, making neuraminidase an attractive target for the development of broad-spectrum inhibitors [6]. Secondly, resistance to neuraminidase inhibitors develops less commonly than to other anti-influenza drugs. Nevertheless, the intensive application of neuraminidase inhibitors for influenza treatment results in a permanently increasing number of drug-resistant strains [7]. Thirdly, in contrast to amantadines, neuraminidase inhibitors are mostly well tolerated in patients under therapy [8]. Finally, neuraminidase protein is a freely accessible target for antiviral molecules with an extracellular mode of action.
The anti-influenza drugs are usually classified according to their target in the viral life-cycle. Such antiviral molecules are particularly used as inhibitors of the following processes: attachment of the virus to host cell receptors, endocytosis and fusion of viral and cell membranes, replication and transcription of the viral genome, synthesis of viral proteins, assembly of the viral progeny and release of the new virions into the outside environment. The following paragraphs are focused on the description of basic classes of influenza virus inhibitors.
Avian influenza A consists of two major glycoproteins which are Hemaglutinin (HA) and Neuraminidase (NA) [9]. HA glycoproteins are more prone to attach to the cell surface sialic acid receptors. There is a difference between host surface receptors on the target cell which is believed to be the possible restrictive factor of avian influenza. HA gene of avian cell binds to Sia2-3Galactosecontaining receptor which is different from human Sia2- 6Galactose containing receptor [10]. Before functioning as a virus it needs post translational cleavage by host proteases [11]. HA followed by NA are important antigenic determinant from which neutralizing antibodies are directed. There are several subtypes of HA and NA. 18 different HA subtypes (H1 to H18) and 11 different NA subtypes (N1 to N11) are found [12]. There is a membrane protein named M2 protein which regulates the internal PH level of the virus. This membrane protein is responsible for un coating the virus during early stages of viral replication [13]. Amantadine and rimantadine block this function. NA catalyze the cleavage of glycosidic linkages to sialic acid on the surface of the viral particle and host cell thus preventing the aggregation and facilitating the release of progeny viruses from the infected cell. Antiviral drugs like Oseltamivir and zanamivir (NA inhibitors) inhibits this important function are the key to the antiviral treatment.
Neuraminidase inhibitors (NAIs) are first-line agents for the treatment and prevention of influenza virus infections. Resistance to the NAIs can be both drug and virus type or subtype specific. The neuraminidase (NA) inhibitors (orally administered oseltamivir and inhaled zanamivir) are currently an important class of antiviral drugs available for the treatment of seasonal and pandemic influenza. Although administration of NA inhibitors may significantly reduce influenza virus transmission, it risks the emergence of drug-resistant variants [14,15]. Several mutations in seasonal or pandemic strains confer resistance to oseltamivir, which is currently the most widely used drug. These mutations are found mainly in the framework of the enzyme and they appear to destabilize drug binding to the target enzyme, thereby reducing viral susceptibility to the treatment [16,17]. Understanding the impact of the H275Y is important for several reasons: (i) it is the most frequent mutation conferring resistance to oseltamivir and (ii) it was detected and transmitted in some viral backgrounds in the absence of antiviral pressure. Indeed, close to 100% of A/ Brisbane/59/ 2007 (H1N1)-like viruses that circulated in 2008– 2009 in Europe and North America were resistant to oseltamivir due to the H275Y NA mutation [18]. These data indicate that, in the appropriate viral background, that is, with the required permissive NA mutation(s), the H275Y mutant can retain ?tness and become more transmissible [19,20]. Based on ferret experiments, many groups have shown that the oseltamivirresistant A (H1N1) pdm09 virus with the H275Y mutation was as virulent as its wild-type counterpart with the exception of a reduced airborne transmission reported in some but not all studies [21-25]. One of the earliest reports of resistance was in summer campers in the USA undergoing oseltamivir prophylaxis. Resistant virus was isolated from the second patient several days after contact with the ?rst. In addition to the H274Y mutation, both viruses had a common I222V (I223V in N1 numbering) mutation, suggesting possible human–human spread [26]. Another case was reported after prophylaxis of a family contact of an infected patient [27]. As the dose of oseltamivir for prophylaxis is only half that for therapy (75 mg once daily), administering subtherapeutic doses when virus replication has already begun could increase the selection of resistant A (H1N1) pdm09 virus.
Transmission
Transmission pattern of avian influenza A from one bird to another is poorly understood because of its complexity, huge number of species among birds and environmental factors. Some experiments have been done to identify the transmission pattern and it shows poorer transmission from infected to susceptible animals [28-30]. Migration process can influence transmission of viruses. Migratory birds can carry pathogens from country to country thereby playing a role distributing influenza viruses.
Materials and Methods: Sequence and Data Source
Data used in this study are obtained using nucleotide BLAST search from publicly available database of National Centre for Biotechnology Information (NCBI). Multiple sequence alignments, editing, assembly of strains were performed in windows platform with the Geneious program version 7.1.3 (trial). Numbers at nodes in the tree indicate Neighbor-Joining bootstraps value generated from 1,000 replicates.
Result and Discussion
In 2010 and 2013 we have selected total 38 strains of Neuraminidase (NA) strain of H1N1 (Figure 1 and Table 1).
| Accession number |
Strain name |
| JN381405 |
A/TN/65229/10 |
| KF559896 |
A/HS/636/13 |
Table 1: List of diverse strains from our analysis of 2010 and 2013 are given below.
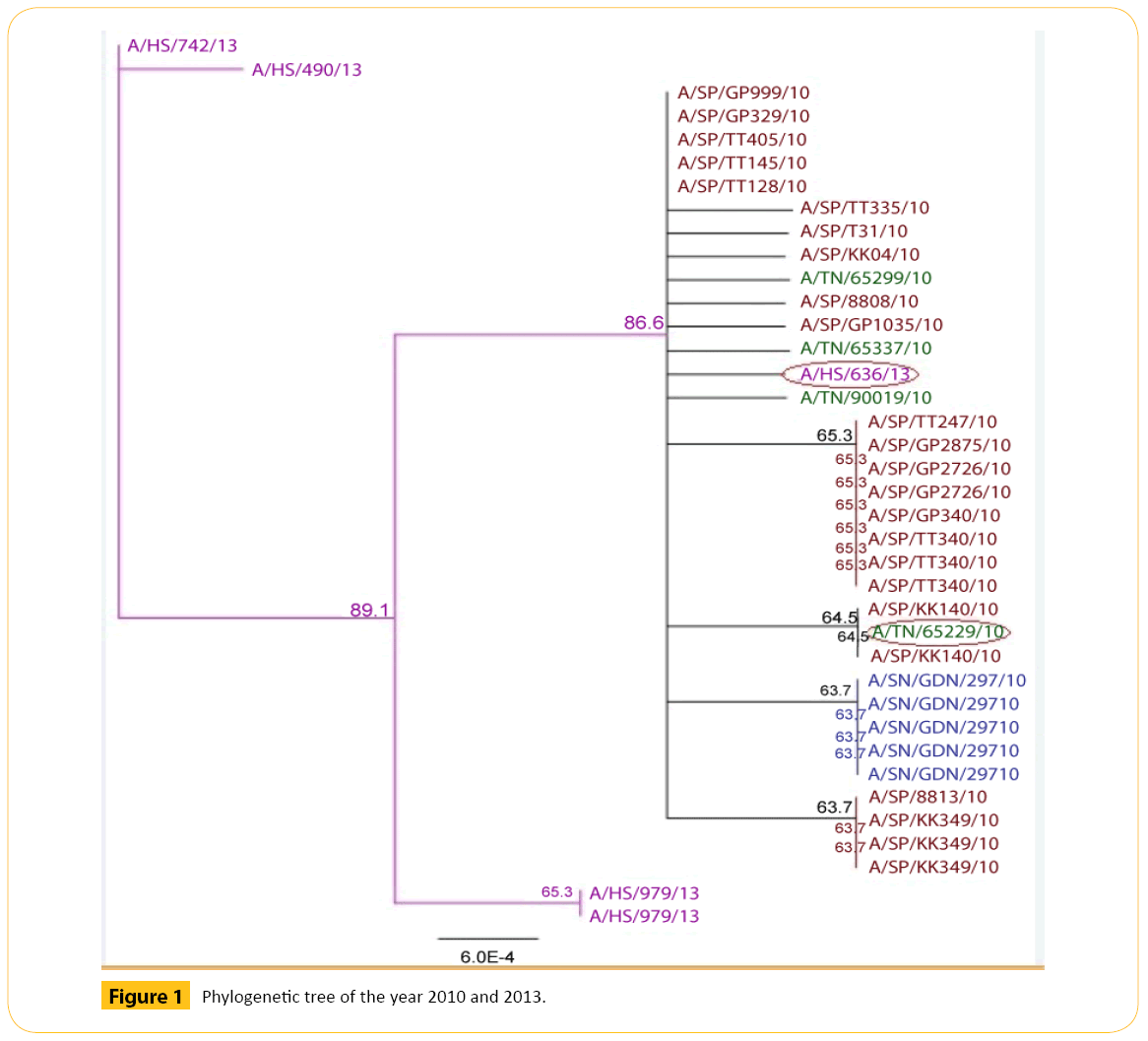
Figure 1: Phylogenetic tree of the year 2010 and 2013.
From two years we have got some strains which seems to diverse from our analysis. We did our literature search but we did not get any information about these diverse strains. So it seems to us that these strains are not responsible for antigenic shift. Neighbor joining method and bootstrap value shows that this diverse strain is showing antigenic drift which may transfer to the other avian in the same country or other different country as well through migratory process.
From the phylogenetic tree of 2010 and 2013 we came to see that all TN like viruses formed a cluster. Among them A/ TN/65229/10 shows 64.5 bootstrap value with A/SP/KK140/10. Number of bootstrap value is showing the similarities between these two strains. Strain A/TN/65229/10may be migrated or may be mutated which can show antigenic drift.A/HS/636/13 shows 86.5 bootstrap values with A/TN/65337/10 and others strains. Number of bootstrap value is showing the similarities between others strains. Strain A/HS/636/13may be migrated or may be mutated which can show antigenic drift.
The molecular mechanisms that enable avian influenza viruses to cross the species barrier and transmit efficiently in humans are incompletely understood. Some experiments have been done to identify the transmission pattern and it shows poorer transmission from infected to susceptible animals [29-31]. Migration process can influence transmission of viruses. Migratory birds can carry pathogens from country to country thereby playing a role distributing influenza viruses. Avian influenza A consists of two major glycoproteins which are Hemagglutinin (HA) and Neuraminidase (NA) [32]. HA glycoproteins are more prone to attach to the cell surface sialic acid receptors. There is a difference between host surface receptors on the target cell which is believed to be the possible restrictive factor of avian influenza.
Human infections are periodic. In some cases these viruses are accompanied by high mortality. As a result they are the major concern about the potential H1N1 as an endemic virus.
The pandemic H1N1 influenza viruses also possess the N1 NA from avian sources [33,34]. Unlike previously circulating viruses, the pandemic H1N1 viruses contained a complex of influenza virus genes of Eurasian and North American swine influenza virus origin [35,36] which were previously derived from reassorted genes of human, swine, and avian lineages. This novel reassortment of HA and NA genes resulted in a virus that was effectively transmitted in humans. In type 1 NA, the major amino acid change causing antiviral resistance is a histidine to tyrosine change at position 274 (H274Y) (Table 2). This change causes an increase of the inhibitor of OSC and PeR [37]. This mutation is the major OSresistance change in influenza A/H1N1 viruses [38].
| Amino acid mutation position with reference |
Short form |
Amino acid in Geneious in reported position |
NAIs |
Asn-295-Ser
[40] |
N295S |
V |
Oseltamivir
Peramivir |
Asn-294-Ser
[38] |
N294S |
C |
Oseltamivir
Peramivir |
His-275-Try
[40] |
H275Y |
A |
Oseltamivir
Peramivir |
His-274-Try
[38] |
H274Y |
N |
Oseltamivir
Peramivir |
Ser-246-Asn
[40] |
S247N |
T |
Oseltamivir
Peramivir |
Ser-246-Asn
[41] |
S246N |
M |
Oseltamivir
Peramivir |
Glu-119-Val
[40] |
E119V |
R |
Oseltamivir
Peramivir |
Ile-222-Arg
[41] |
I222R |
S |
Oseltamivir
Zanamivir |
Table 2: Summary of some reported position which can be responsible for N1 (H1N1) neuraminidase mutations causing reduced sensitivity to NAIs..
Before functioning as a virus it needs post translational cleavage by host proteases [12]. HA followed by NA are important antigenic determinant from which neutralizing antibodies are directed. There are several subtypes of HA and NA. 18 different HA subtypes (H1 to H18) and 11 different NA subtypes (N1 to N11) are found [39].
Data used in this study are obtained inside using nucleotide BLAST search from publicly available database of National Centre for Biotechnology Information (NCBI). Multiple sequence alignments, editing, assembly of strains were performed in windows platform with the Geneious program version 7.1.3 (trial).
In this study we will analyze some swine neuraminidase (H1N1) of different years. Analysis includes building nucleotide sequence and translating them into amino acid sequence. Then we will study amino acid positions with respect to some reported mutation to see the genetic pattern.
We found some avian amino acid position N295S, N294S, H275Y, H274Y, S247N, S246N, E119V and I222Rare specific reported position which can be responsible for N1 (H1N1) neuraminidase mutations causing reduced sensitivity to NAIs [38,40,41]. In our swine H1N1 analysis we did not find the exact location where reported mutations are occurred. But we found similar amino acid near the reported mutated position. We have analyzed around (before and after the mutation point) twenty positions with respect to the reported mutation point.
Correlation of Reported NAIs resistance specific avian amino acid position with our experimental strain, and their mutation pattern (exact and around twenty positions): Original Amino Acid analysis (Table 2).
In case of N295S (Asn-295-Ser) reported position, we found Valine (V) in our software (Geneious).
There is no mutation here. We found amino acid Asparagine (N) in one positions (277) which is located within twenty positions before 295. There is no mutation here. We found amino acid Asparagine (N) in four positions (299, 304, 311 and 313) which is located within twenty positions after 295. There are no mutations here (Table 3).
| Reported AA with mutation point |
AA in Geneious in reported position |
Exact AA which matches with the reported data with respect to specific mutation point (before 20 positions) |
Exact AA which matches with the reported data with respect to specific mutation point (after 20 positions) |
| N295S |
V295 |
N277 |
N299
N304
N311
N313 |
| N294S |
C294 |
N277 |
N299
N304
N311
N313 |
| H275Y |
A275 |
|
H279Y(1)
Electrically charged Positive to Polar |
| H274Y |
N274 |
|
H279Y(1)
Electrically charged Positive to Polar |
| S247N |
T247 |
S241
S233 |
S251N(1)
Polar to Polar
S256 |
| S246N |
M246 |
S241
S233 |
S251N(1)
Polar to Polar
S256 |
| E119V |
R119 |
|
E120K(1)
Electrically charged Negative to Positive
E129 |
| I222R |
S222 |
I220
I216
I215 |
I227T(1)
Nonpolar to Polar |
Table 3: Analysis of our study data with respect to reported mutation point causing reduced sensitivity to NAIs of swine H1N1.
In case of N294S (Asn-294-Ser) reported position, we found Cysteine (C) in our software (Geneious).
There is no mutation here. We found amino acid Asparagine (N) in one positions (277) which is located within twenty positions before 294. There is no mutation here. We found amino acid Asparagine (N) in four positions (299, 304, 311 and 313) which is located within twenty positions after 294. There are no mutations here.
In case of H275Y (His-275-Try) reported position, we found Alanine (A) in our software (Geneious).
There is no mutation here. We found amino acid Histidine (H) in one positions (279) which is located within twenty positions after 275. There is one mutation here from Histidine (H) to Tyrosine (Y) in one strain. We found H279Y mutation which is indicate that there is polarity changed from electrically charged Positive to polar as Histidine (H) is electrically charged Positive and Tyrosine (Y) is polar.
In case of H274Y (His-274-Try) reported position, we found Asparagine (N) in our software (Geneious).
There is no mutation here. We found amino acid Histidine (H) in one positions (279) which is located within twenty positions after 274. There is one mutation here from Histidine (H) to Tyrosine (Y) in one strain. We found H279Y mutation which is indicate that there is polarity changed from electrically charged Positive to polar as Histidine (H) is electrically charged Positive and Tyrosine (Y) is polar.
In case of S247N (Ser-247-Asn) reported position, we found Threonine (T) in our software (Geneious).
There is no mutation here. We found amino acid Serine (S) in two positions (233 and 241) which is located within twenty positions before 247. There are no mutations here. We found amino acid Serine (S) in two positions (251 and 256) which is located within twenty positions after 247. There is one mutation here from Serine (S) to Asparagine (N) in one strain. We found S251N mutation which is indicates that there is polarity changed from polar to polar as Serine (S) is polar and Asparagine (N) is polar.
In case of S246N (Ser-246-Asn) reported position, we found Methionine (M) in our software (Geneious).
There is no mutation here. We found amino acid Serine (S) in two positions (233 and 241) which is located within twenty positions before 246. There are no mutations here. We found amino acid Serine (S) in two positions (251 and 256) which is located within twenty positions after 246.There is one mutation here from Serine (S) to Asparagine (N) in one strain. We found S251N mutation which is indicates that there is polarity changed from polar to polar as Serine (S) is polar and Asparagine (N) is polar.
In case of E119V (Glu-119-Val) reported position, we found Arginine (R) in our software (Geneious). There is no mutation here. We found amino acid Glutamic acid (E) in two positions (120 and 129) which is located within twenty positions after 119. There is one mutation here from Glutamic acid (E) to Lysine (K) in one strain. We found E120K mutation which is indicates that there is polarity changed from electrically charged negative to Positive as Glutamic acid (E) is electrically charged negative and Lysine (K) is electrically charged Positive.
In case of I222R (Ile-222-Arg) reported position, we found Serine (S) in our software (Geneious).
There is no mutation here. We found amino acid Isoleucine (I) in three positions (225, 216 and 220) which is located within twenty positions before 222. There are no mutations here. We found amino acid Isoleucine (I) in one position (227) which is located within twenty positions after 222. There is one mutation here from Isoleucine (I) to Threonine (T) in one strain. We found I227T mutation which is indicates that there is polarity changed from non-polar to polar as Isoleucine (I) is non-polar and Threonine (T) is polar.
In case of N295S (Asn-295-Ser) reported position, we found Valine (V) in our software (Geneious).
There is no mutation here. We found amino acid Serine (S) in three positions (284, 289 and 290) which is located within twenty positions before 295. There are two mutations here from Serine (S) to Asparagine (N) in two strains and from Serine (S) to Glycine (G) in one strain. We found S290N and S290G mutations which are indicate that there is polarity changed from polar to polar and from polar to non-polar as Serine (S), Asparagine (N) is polar and Glycine (G) is non-polar. We found amino acid Serine (S) in two positions (303 and 309) which is located within twenty positions after 295. There are no mutations here (Table 4).
| Reported AA with mutation point |
AA in Geneious in reported position |
Exact AA which matches with the reported data with respect to specific mutation point (before 20 positions) |
Exact AA which matches with the reported data with respect to specific mutation point (after 20 positions) |
| N295S |
V295 |
S290N(2)
Polar to Polar
S290G(1)
Polar to Nonpolar
S289
S284 |
S303
S309 |
| N294S |
C294 |
S290N(2)
Polar to Polar
S290G(1)
Polar to Nonpolar
S289
S284 |
S303
S309 |
| H275Y |
A275 |
Y257 |
Y278
Y280
Y286 |
| H274Y |
N274 |
Y257 |
Y278
Y280
Y286 |
| S247N |
T247 |
N239 |
|
| S246N |
M246 |
N239
N226S(3)
Polar to Polar |
|
| E119V |
R119 |
V117I(3)
Nonpolar to Nonpolar
V115 |
|
| I222R |
S222 |
|
R224
R229 |
Table 4: Correlation of Reported NAIs resistance specific avian amino acid position with our experimental strain, and their mutation pattern (exact and around twenty positions): Mutated Amino Acid analysis.
In case of N294S (Asn-294-Ser) reported position, we found Cysteine(C) in our software (Geneious).
There is no mutation here. We found amino acid Serine (S) in three positions (284, 289 and 290) which is located within twenty positions before 294. There are two mutations here from Serine (S) to Asparagine (N) in two strains and from Serine (S) to Glycine (G) in one strain. We found S290N and S290G mutations which are indicate that there is polarity changed from polar to polar and from polar to non-polar as Serine (S), Asparagine (N) is polar and Glycine (G) is non-polar. We found amino acid Serine (S) in two positions (303 and 309) which is located within twenty positions after 294. There are no mutations here.
In case of H275Y (His-275-Try) reported position, we found Alanine (A) in our software (Geneious).
There is no mutation here. We found amino acid Tyrosine (Y) in one positions (257) which is located within twenty positions before 275. There is no mutation here. We found amino acid Tyrosine (Y) in three positions (278, 280 and 286) which are located within twenty positions after 275. There are no mutations here.
In case of H274Y (His-274-Try) reported position, we found Alanine (A) in our software (Geneious).
There is no mutation here. We found amino acid Tyrosine (Y) in one positions (257) which is located within twenty positions before 274. There is no mutation here. We found amino acid Tyrosine (Y) in three positions (278, 280 and 286) which are located within twenty positions after 274. There are no mutations here.
In case of S247N (Ser-247-Asn) reported position, we found Threonine (T) in our software (Geneious).
There is no mutation here. We found amino acid Asparagine (N) in one position (239) which is located within twenty positions before 247. There are no mutations here.
In case of S246N (Ser-246-Asn) reported position, we found Threonine (T) in our software (Geneious).
There is no mutation here. We found amino acid Asparagine (N) in one position (239) which is located within twenty positions before 246. There are no mutations here.
In case of E119V (Glu-119-Val) reported position, we found Arginine (R) in our software (Geneious). There is no mutation here. We found amino acid Valine (V) in two positions (115 and 117) which is located within twenty positions before 119. There is one mutation here from Valine (V) to Isoleucine (I) in three strains. We found V117I mutation which is indicates that there is polarity changed fromnon-polar to polar as Valine (V) is non-polar and Isoleucine (I) is polar.
In case of I222R (Ile-222-Arg) reported position, we found Serine (S) in our software (Geneious).There is no mutation here. We found amino acid Arginine (R) in two positions (224 and 229) which is located within twenty positions after 222. There are no mutations here.
Conclusion
Resistance to the NAIs can be both drug and virus type or subtype speci?c. A summary of the effects of mutations referred to in this review is presented in Table 2. In the N1 subtype, the most frequently encountered mutation is the H275Y that confers highly reduced inhibition to oseltamivir, moderate crossresistance to the investigational agent peramivir and susceptibility to zanamivir. Various amino acid changes at residue 222 (I ?R) or 223 (I ?R/V) can also confer reduced inhibition to oseltamivir and/or to zanamivir.In our Geneiouswe found some similar amino acids near the reported mutated positions that may cause reduce sensitivity to NAIs.
The global outbreak of the human pandemic influenza A virus (H1N1) has caused a significant alertness in the general public [34]. Analysis of the genetic material allows a prediction of the new influenza virus characteristics including its capacity to spread the infection, degree of virulence, and drug resistance. In our article we analyzed phylogenetic tree and found some diverse strains those may be migrated or may be mutated which can show antigenic drift.
At present, it is clear that mutation conferring resistance to the currently approved antiviral drugs is a growing problem, so it is very important to continue research in various areas that would allow better resolution of the problem: First, the knowledge of mutation’s type that allows virus avoids the action of drugs, will allow an understanding as to how these mutations arise and how we must avoid them. Also, this knowledge, will allow a better design of new drugs.
Conflict of Interest: None
6750
References
- Inoue M, Barkham T, Leo YS (2010) Emergence of oseltamivir-resistant pandemic (H1N1) 2009 virus within 48 hours. Emerg Infect Dis 16:1633-1636.
- Valinotto LE, Diez RA, Barrero PR (2010) Emergence of intratreatment resistance to oseltamivir in pandemic influenza A H1N1 2009 virus.AntivirTher 15:923-927.
- Renaud C, Pergam SA, Polyak C (2010) Early emergence of an H275Y mutation in a hematopoietic cell transplant recipient treated with intravenous peramivir. Transpl Infect Dis 12:513-517.
- Matrosovich MN, Matrosovich TY, Gray T, Roberts NA, Klenk HD (2004) Neuraminidase is important for the initiation of influenza virus infection in human airway epithelium. Journal of Virology 78: 12665-12667.
- Suzuki T, Takahashi T, Guo CT, Hidari KIPJ, Miyamoto D, et al. (2005) Sialidase activity of influenza A virus in an endocytic pathway enhances viral replication. Journal of Virology 79: 11705-11715.
- Yen HL, Hoffmann E, Taylor G, Scholtissek C, Monto AS, et al. (2006) Importance of neuraminidase active-site residues to the neuraminidase inhibitor resistance of influenza viruses.Journal of Virology 80: 8787-8795.
- Garcia J, Sovero M, Torres AL, Gomez J, Douce R, et al. (2009) Antiviral resistance in influenza viruses circulating in Central and South America based on the detection of established genetic markers. Influenza and Other Respiratory Viruses 3: 69-74.
- Cao B, Wang DY, Yu XM, We LQ, Pu ZH, et al. (2012) An uncontrolled openlabel, multicenter study to monitor the antiviral activity and safety of inhaled zanamivir (as Rotadisk via Diskhaler device) among Chinese adolescents and adults with influenza-like illness. Chinese Medical Journal 125: 3002-3007.
- Ito T, Goto H, Yamamoto E, Tanaka H, Takeuchi M, et al. (2001) Generation of a highly pathogenic avian influenza A virus from an avirulent field isolate by passaging in chickens. J Virol75:4439-4443.
- Matrosovich M, Zhou N, Kawaoka Y, Webster R (1999) The Surface Glycoproteins of H5 Influenza Viruses Isolated from Humans, Chickens, and Wild Aquatic Birds Have Distinguishable Properties. Journal of Virology 73: 1146-1155.
- Rott R (1992) The pathogenic determinant of influenza virus.Veterinary Microbiology 33: 303-310.
- Tong S, Zhu X, Li Y, Shi M, Zhang J, et al. (2013) New World Bats Harbor Diverse Influenza A Viruses. PLoS Pathogens 9: e1003657.
- Jing X, MaC,Ohigash Y, Oliveira FA,Jardetzky TS, et al. (2008) Functional studies indicate amantadine binds to the pore of the influenza A virus M2 proton-selective ion channel. Proc Natl Acad Sci USA 105: 10967-10972.
- Mendel DB, Roberts NA (1998) In-vitro and in-vivo efficacy of influenza neuraminidase inhibitors. CurrOpin Infect Dis 11:727-732.
- Dgxncgh SK, Mitamura K, Sakai TY, Goto H, Sugaya N, et al. (2003) High frequency of resistant viruses harboring different mutations in amantadine-treated children with influenza. J Infect Dis 188:57-61.
- Russell RJ, Haire LF, Stevens DJ, Collins PJ, Lin YP, et al. (2006) The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design. Nature 443: 45-49.
- Van der Vries E, Schutten M, Boucher CA (2011)The potential for multidrug-resistant influenza. Current Opinion Infectious Diseases 24: 599-604.
- Dharan NJ, Gubareva LV, Meyer JJ (2009) Infections with oseltamivirresistant influenza A(H1N1) virus in the United States. JAMA 301:1034-1041.
- Bloom JD, Gong LI, Baltimore D (2010) Permissive secondary mutations enable the evolution of influenza oseltamivir resistance. Science 328:1272-1275.
- Bloom JD, Nayak JS, Baltimore D (2011) A computational-experimental approach identifies mutations that enhance surface expression of an oseltamivir-resistant influenza neuraminidase. PLoS ONE 6: e22201.
- Hamelin ME, Baz M, Abed Y (2010) Oseltamivir-resistant pandemic A/ H1N1 virus is as virulent as its wild-type counterpart in mice and ferrets. PLoSPathog 6:e1001015.
- Hamelin ME, Baz M, Bouhy X (2011) Reduced airborne transmission of oseltamivir-resistant pandemic A/H1N1 virus in ferrets. AntivirTher 16:775-779.
- Duan S, Boltz DA, Seiler P (2010) Oseltamivir-resistant pandemic H1N1/ 2009 influenza virus possesses lower transmissibility and fitness in ferrets. PLoSPathog 6:e1001022.
- Kiso M, Shinya K, Shimojima M (2010) Characterization of oseltamivirresistant 2009 H1N1 pandemic influenza A viruses. PLoSPathog 6:e1001079.
- Seibert CW, Kaminski M, Philipp J (2010) Oseltamivir-resistant variants of the 2009 pandemic H1N1 influenza A virus are not attenuated in the guinea pig and ferret transmission models. J Virol 84:11219-11226.
- CDC (2009) Oseltamivir-resistant 2009 pandemic influenza A (H1N1) virus infection in two summer campers receiving prophylaxis-North Carolina. MMWR Morb Mortal Wkly Rep 58:969-972.
- Baz M, Abed Y, Papenburg J (2009) Emergence of oseltamivir-resistant pandemic H1N1 virus during prophylaxis. N Engl J Med 361:2296-2297.
- Narayan O, Lang G Rouse ST (1969) A new influenza A virus infection in turkeys. IV Experimental susceptibility of domestic birds to virus strain ty/Ontario/7732/1966.Archivfur diegesamte Virusforschung 26: 149-165.
- Alexander DJ, Allan WH, Parsons D, Parsons G (1978) The pathogenicity of four avian influenza viruses for fowls, turkeys and ducks.Research in Veterinary Science 24: 242-247.
- Westbury HA, Turner AJ, Kovesdy L (1979) The pathogenicity of three Australian fowl plague viruses for chickens, turkeys and ducks. Veterinary Microbiology4:223-234.
- Garten RJ, Davis CT, Russell CA, Shu B, Lindstrom S, et al. (2001) Generation of a highly pathogenic avian influenza A virus from an avirulent field isolate by passaging in chickens. J Virol75:4439-4443.
- Gambotto A, Barratt-Boyes SM, de Jong MD, Neumann G, Kawaoka Y (2008) Human infection with highly pathogenic H5N1 influenza virus. Lancet371:1464-1475.
- Cohen J (2009) New details on virus’s promiscuous past. Science 324:1127.
- Dawood FS, Jain S, Finelli L, Shaw MW, Lindstrom S(2009) Emergence of a novel swine-origin influenza A (H1N1) virusin humans. N. Engl. J. Med 360: 2605-2615.
- Bridges MS, Jernigan DB, Uyeki TM, Smith DJ, Klimov AI, et al. (2009) Antigenic and genetic characteristics of swine-origin 2009 A (H1N1) influenza viruses circulating in humans. Science 325:197-201.
- Smith GJD, Dhanasekaran V, Bahl J, Lycett SJ, Worobey M, et al. (2009) Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 459:1122-1125.
- Samson M, Pizzorno A, Abed Y, Boivin g (2013) Influenza virus resistance to neuraminidase inhibitors. Antiviral Res 98: 174-185.
- Le QM, Kiso M, Someya K, Sakai YT, Nguyen TH, et al. (2005) Avian flu: isolation of drugresistant H5N1 virus. Nature 437: 1108.
- Shinya K, Ebina M, Yamada S, Ono M, Kasai N, et al. (2006) flu: influenza virus receptors in the human airway. Nature440:435-436.
- Pizzorno A, Abed Y, Boivin G 92011) In?uenza drug resistance.SeminRespirCrit Care Med 32:409-422.
- Nguyen HT, Fry AM, Loveless PA, Klimov AI, Gubareva LV (2010) Recovery of a multidrugresistant strain of pandemic influenza A 2009 (H1N1) virus carrying a dual H275Y/I223R mutation from a child after prolonged treatment with oseltamivir. Clin Infect Dis 51: 983- 984.






