Isolation and In Vitro Evaluation of Bacteriophage against Methicillin-Resistant Staphylococcus aureus (MRSA) from Burn wounds
Keywords
MRSA; Burnt wound; Bacteriophage; Lytic activity; Hospital sewage
Introduction
The increasing appearance of multi-drug resistant (MDR) microorganisms in clinics is a rising serious threat to human health [1]. Most recently, an estimated 2.5 million people acquire antibiotic-resistant infections every year in Europe and USA leading to approximately 50,000 deaths [2].
Staphylococcus aureus is a common commensal bacterium which is an opportunistic pathogen capable of causing a variety of diseases ranging from food poisoning to fatal infections like endocarditis, pneumonia, osteomyelitis and toxic shock syndrome. It is also a leading cause of infections associated with catheters or devices [3-6].
Methicillin-resistant Staphylococcus aureus (MRSA) is an MDR (multi-drug resistant) organism that is resistant to every antibiotic except for Vancomycin. It was initially detected during early 1960's in United Kingdom and is now regarded as a major hospital acquired pathogen throughout the world [7,8]. Also, community-acquired MRSA infections have increasingly been identified within the past two decades [5]. This microorganism has been declared an international concern by the World Health Organization [9,10].
In addition to the increasing prevalence of MRSA, the emergence of vancomycin-resistant or vancomycinintermediate Staphylococcus aureus (VRSA or VISA) has caused serious concern [7,11]. These infections have different manifestations and include infection of the epidermis, dermis, superficial fascia, subcutaneous tissues and muscles. More severe SSTIs include deeper soft tissues such as infectious cellulitis, ulcer or wound site infection, surgical site infection, major abscesses, skin ulcers, and diabetic foot ulcers [12,13]. MRSA is also one of the most common nosocomial pathogens infecting burn wounds [14]. It is estimated that 70% of the mortality in burn unit is related to infections. Thus, management of burn patients is a significant problem because of outbreaks of infection in burn units and the presence of many multi drug resistant strains. Additionally Hospital burns units are a main reservoir for MRSA with a potential for rapid dissemination in hospital environment [15]. Shahsavan et al. [14] among Staphylococcus aureus strains isolated from burnt patients in Tehran reported that the most of MRSA isolates were MDR that showed resistance to β-lactams, macrolides, tetracyclines and aminoglycosides.
As a result, therapeutic strategies to counter clinical infectious caused by these bacteria has become limited [2]. Therefore, it is critical that alternative antibacterial agents and programs are developed [2]. Moreover, due to rapid acquisition of resistance to the new antibiotics and rising production costs, there has been little incentive to develop new antibacterial agents [2,11,16].
A strategy against MRSA infections and to provide a new solution against the threat of MDR infections, phage therapy has attracted a great deal of attention [17]. This approach uses bacterial viruses (phages) which can specifically attack pathogenic bacteria and kill them [18]. Phage therapy has the potential of being highly specific against only the species and even strain responsible for an infection [3,19,20]. Moreover, their remarkable specificity prevents them from affecting human cells, microbial composition of body microbiota and inducing antimicrobial resistance in different bacterial species [20,21].
In this study, a specific bacteriophage was isolated from hospital sewage which was effective against MRSA strains from burnt patient. The morphological feature of the phage, its nucleic acid composition, host range, latent time and burst size were characterized.
Materials and Methods
Isolation and characterization of bacterial strains
Thirty S. aureus strains isolated from burnt patients hospitalized at Motahari Hospital in Tehran, Iran plus reference strain of MRSA (ATCC 43300) were used as hosts for bacteriophage isolation and propagation from the wastewater samples. Strains were confirmed as S. aureus by gram stain, coagulase, catalase, oxidase and DNase production as well as mannitol fermentation and PCR amplification of protein A gene (spa).
Kirby-Bauer disk diffusion tests were performed to confirm its resistance to penicillin, methicillin, and oxacillin as recommended by Clinical and Laboratory Standards Institute (CLSI) [21]. Colonies from an overnight pure culture of each isolate were emulsified in sterile normal saline with the turbidity adjusted to 0.5 McFarland standards.
The bacterial suspension was uniformly streaked on Mueller Hinton agar and left to dry for 3-5 min. Then antibiotic disks, placed on the agar surface and incubated at 35°C for 18-24 hours [22]. Additionally PCR assays were carried out for mecA gene. Bacteria from LBA (Luria Bertani Agar, Thermo Scientific Oxoid) plates were grown at 37°C for 4 h to reach the exponential phase with constant shaking (180 rpm) in LB broth(Luria Broth, Thermo Scientific Oxoid).
Wastewater sample collection for bacteriophage isolation
Several wastewater samples were taken from untreated sewage pond of Motahari Hospital and screened as a source of bacteriophages. The samples were centrifuged at 2,500×g for 20 min to separate bacterial cell and debris. The supernatant was filtered through 0.45 μm filters, transferred to a clean tube and stored at 4°C.
Isolation and purification of bacteriophages
For phage isolation, 10 ml exponential phase culture of a clinical and standard S. aureus was mixed with 10 ml of fresh LB broth (2X) and 10 ml of filtered sewage sample, and incubated overnight at 37°C with shaking at 80 rpm. The culture was then centrifuged for 20 min at 2,500×g, to remove bacteria, and the supernatant was filtered through 0.22 μm pore size Millipore filter (for phage amplification the simultaneous culturing step was repeated for three or more times).This lysate (supernatant) was examined for lytic phage in plaque assay by double layer agar(DLA) method.
Double layer agar (DLA)
One ml of phage lysate was mixed with500 μl of a stationary phase culture of host S. aureus after10 min incubation at 37°C, 5 ml of LB top agar (4 mM CaCl2, 4 mM MgSO4, 0.7% (w/v) agar, at 45°C) was added, mixed and overlaid on to fresh LB agar plates(1.5 % (w/v) agar )prepared before. After solidifying, plates were incubated overnight at 37°C until lysis zones appeared.
Purification of bacteriophage
Single plaques from each plate were picked by sterile Pasteur pipette and placed in a tube containing 1 ml of LB broth (45°C).One ml of stationary phase culture of host bacteria was added to each tube and incubated for 24 h at 37°C and 80 rpm. Next day, the lysate were screened for presence of plaques as describe above. Purification was carried out by three serial single-plaque isolations.
Determination of bacteriophage titer
Ten sterile tubes containing 900 μl Luria Bertani (LB) broth were numbered from 10-1 to 10-10. One hundred μl of phage lysate was added to the first tube (10-1) mixed well and 100 μl was transferred to the second tube (10-2) in the series (10-1 to 10-10). The same pattern was used to make serial dilutions. Then, 100 μl of an exponential phase culture of the host MRSA were added to each tube and mixed with the 100 μl of each dilution of phage lysate (10-5 to 10-9). Tubes were incubated for 10 min at 37°C for phages adsorption.
Five tubes containing 3 ml LB soft agar (0.4% agar, 4 to 10 mM CaCl2 and MgSO4) at 45°C were numbered from 10-5 to 10-9.The phage and bacterial suspensions were added to soft agar tubes and after mixing were overlaid on to plates containing LB agar (1.5% agar).Plates were incubated overnight at 37°C and the resulting plaques were counted. PFU/ml was equal to number of plaques/dilution × volume of diluted phage added to each plate.
Host range determination
Thirty clinical MRSA isolates, recovered from burnt patient, were used to determine host range of the bacteriophage isolated in this study. To determine MSSA (methicillinsusceptible S.aureus) strains susceptibility to phage-mediated lysis, 30 strains from burnt patient were also examined. Bacterial strain susceptibilities were detected by the spot test method. Briefly, bacterial strains were incubated 4 h in LB broth at 37°C and 180 rpm (OD 600 nm=0.4-0.6). Then 500 μl of each bacterial culture was added to 6 ml of 0.4% LB soft agar (45°C) and poured onto LB agar plates. The plates were left to dry for 10 minutes. Subsequently, 10 μl of the phage lysate (109 PFU/ml) was spotted on lawns of different bacterial strains and incubated overnight at 37°C for formation of lysis zone. Lytic activity of isolated phage was also examined on Staphylococcus epidermidis, Enterococcus faecalis and Enterococcus fasium strains.
Bacteriophage storage
Phage lysate was filtered and precipitated by adding polyethylene glycol 8,000 (BIO BASIC CANADA INC.Cat. #: PB0433)) and NaCl (MERK, EMSURE® ACS, ISO, Reag. PhEur, Germany) to final concentrations of 10% and 1 M, respectively followed by incubation at 4°C for 18 h. The lysatewere centrifuged at 10,000×g for 20 min. Phage pellet were resuspended in SM buffer (100 mMNaCl, 10 mM MgSO4, 10 mM Tris-HCl [pH 7.5] with 50% (vol/vol) glycerol and stored at 80°C for long-term use. For Short-term use, prepared stocks were stored at 4°C.
Electron microscopy
Ten μl polyethylene glycol precipitated phage particles were spotted onto a carbon-coated copper acid grid for 3-5 minutes and then blotted with filter paper and stained with 1% (w/v) uranyl acetate(pH 7).It was examined by Zeiss LEO 906 transmission electron microscope (Carl Zeiss LEO EM 906 E, Germany) at an accelerating voltage of 100 kV.
Naming the isolated bacteriophage
We named our isolated phage to vB_StuM_MH-1 according to the newly proposed naming system vBStuP/M/S MHno, where vB=bacterial virus; Stu=abbreviation for genus/species of the host; P=podovirus, M=myovirus, S=siphovirus; MHno=name and number of phage. The latter part of the name (MH-1) will be the phage’s common name [23].
One-step growth curve
A mid-exponential-phase culture (30 ml) of S. aureus (OD600 nm=0.4 to 0.5) was harvested by centrifugation and re-suspended in 7.5 ml of fresh LB broth. Phage lysates (109 PFU/ ml) were added at an MOI of 0.0005 and incubated at 37°C for 15 min for phage adsorption. The mixture was then centrifuged at 10,000×g for 10 min to remove free phage particles. Pellet was re-suspended in 10 ml of LB broth and incubated at 37°C. Samples were removed at 10 min intervals for 2 h. The samples were immediately diluted 10 fold and plated for phage titration using DLA method. The next day, plaques were counted and PFU/ml was calculated as mentioned above.
Isolation of bacteriophage genome
Extraction of phage genomic DNA was done according to the Martha RJ and Clokie A M method with slight modification [24]. Briefly 10 ml of phage lysates (with 10% PEG 8000, and 1 M concentration of NaCl). A mixture of 5 μl of 1 mg/ml DNase I and 2 μl of 12.5 mg/ml RNase A was added to the tube and incubated for 30 min at 37°C. Afterward, 12 μl of 20% SDS and 5 μl of 10 mg/ml proteinase K was added to the mixture and incubated for 30 min at 37°C. Extraction was carried out with 0.5 ml of phenol: chloroform: isoamyl alcohol (25:24:1).
The mixture was spun for 5 min at 15,000×g for phase separation. Supernatant was transferred into a fresh tube and extracted once with 0.5 ml chloroform: isoamylalcohol (24:1) fallowed by centrifugation for 5 min at 8,000×g. The supernatant was transferred into a fresh 2 ml Eppendorf tube and 500 μl of 100% isopropanol and 45 μl of 3 M sodium acetate (pH 5.2) were added.
DNA was left for 30 min to precipitate at room temperature. Following centrifugation for 20 min at 14,000×g, the DNA pellet was washed twice with 70% ethanol and let to dry. Dried DNA pellet was re-suspended in 50 μl distilled water. The phage genomic DNA concentrations and quality was determine using a NanoDrop spectrophotometer (Nanodrop One C, Thermo Fisher Scientific, Assembled in USA), following the instructions provided by the manufacture. Ten microliter of genomic DNA was resolved on 0.1% agarose gel electrophoresis with DNA molecular weight marker λ-Hind III digest and 10 Kb plus DNA size marker (Takara).
Restriction digestion of phage DNA
The DNA of isolated bacteriophage was digested with Rnase A, Dnase I and 6 restriction endonucleases (EcoRV, EcoRI, HaeIII, XbaI, SmaI, SacII, EcoRV) purchased from Thermo Scientific (EU) Lithuania for 5-16 hours. After the enzymatic digestion, restriction fragments were separated by electrophoresis in a 1% agarose gel containing safe stain (SMO Bio-FluoroVue Nucleic Acid Gel Stain (Pre-Stain) in TBE buffer (Tris-boricacid-EDTA), at 90 V in a peQ Lab agarose gel electrophoresis system (peQLAB, E0303, Taiwan).Gene Ruler 10 Kb plus DNA Ladder and λ-Hind III digest DNA molecular weight marker (Thermo Fisher Scientific) was used as a size markers. The gel visualized under gel documentation system (VILBER LOURMAT, E-Box CX5.TS, France). Restriction digestions were performed in triplicate.
Results
Bacteria and bactriophage isolation
Thirty MRSA isolates recovered from burnt patient were confirmed by biochemical and molecular tests and chosen for hosts range determination. Three of the isolates and an MRSA standard strain were selected for phage isolation. A total of six phages were isolated. Phage MH-1 was chosen for sensitivity determination and continuing the study.
Host range analysis
For assessment of host range of the lytic phage, a wide spectrum of lysis actively was evaluated. Of the 30 MRSA and 30 MSSA isolates, 27 strains (90%) and 26 strains (86.6%) were sensitive to the isolated phage respectively and formed the zone of lysis in the spot test (Figure 1). The lytic activity of isolated phage was also examined on 3 other bacterial species. All of the S. epidermidis, E. faecalis and E. fasium strains were found to be resistant to our isolated phage (Table 1).

Figure 1: Spot test.
| Bacterial strain |
Number of isolates |
Plaque Formation |
Reference |
| MRSA |
30 |
27 from 30 |
Clinical isolates |
| MSSA |
30 |
26 from 30 |
Clinical isolates |
| S. epidermidis |
30 |
not susceptible to phage MH-1 |
Clinical isolates |
| E. faecalis |
6 |
not susceptible to phage MH-1 |
Clinical isolates |
| E. fasium |
30 |
not susceptible to phage MH-1 |
Clinical isolates |
Table 1. Bacterial strains used for determination of isolated phage host range
Morphology of the lytic bacteriophage
Designation of morphological characteristics by electron microscopy revealed that this phage is a member of the myoviridea family with contractile, long and relatively thick tail and isomeric large head with about 220 and 100 nm in diameter respectively (Figure 2).

Figure 2: TEM image of phage MH-1 belonging to the family Myoviridae.
A indicate the noncontracted and B contracted tails, respectively. The phages were negatively stained with 1% (wt/ vol) uranyl acetate and observed using Zeiss LEO 906 transmission electron microscope (Carl Zeiss LEO EM 906 E, Germany) at an accelerating voltage of 100 kV.
Determination of bacteriophage titer
Phage titer was ascertained by serial dilution and DLA method. Plaques were counted after overnight incubation at 37°C and titer of the isolated phage was determined to be 1×109 PFU/ml (Figure 3).
Figure 3: The titer of bacteriophage MH-1 assessed with DLA method.
Latent time and phage burst size
To determine the latent time and burst size of the phage MH-1, one-step growth curve analysis was performed. From the analysis of this curve latent period was estimated to be about 20 min. The burst size was 190 PFU per infected cell (Figure 4).
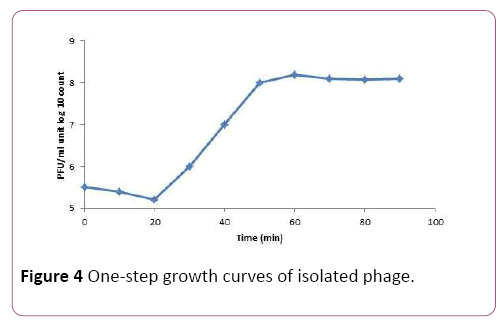
Figure 4: One-step growth curves of isolated phage.
Isolation of genomic DNA of bacteriophages Phage genomic DNA was extracted with phenol: chloroform method and resolved on 0.1% agarose gel electrophoresis. The phage genome was totally digested by DNase I but not by RNase A. Genomic DNA of the phage was digested by the EcoRI, HaeIII, XbaI, SacII and EcoRV restriction enzymes. However, the phage genome seemed to be resistant to digestion by the SmaI restriction enzyme. Also, digestion profiles of XbaI restriction enzyme show that the genome of isolated phage was <∼43 kbp (Figure 5).
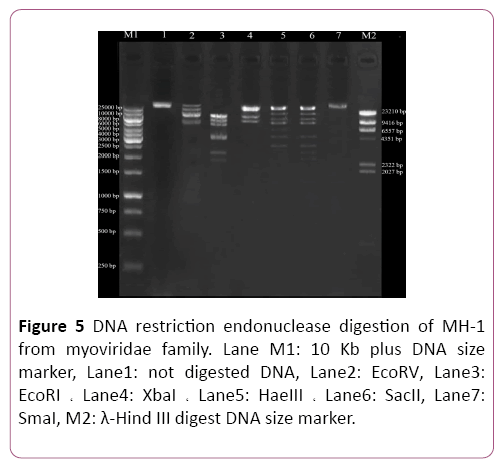
Figure 5: DNA restriction endonuclease digestion of MH-1 from myoviridae family. Lane M1: 10 Kb plus DNA size marker, Lane1: not digested DNA, Lane2: EcoRV, Lane3: EcoRI ? Lane4: XbaI ? Lane5: HaeIII ? Lane6: SacII, Lane7: SmaI, M2: λ-Hind III digest DNA size marker.
Discussion
One of the greatly increasing causes lead to mortality and morbidity in burn patients is colonization of Methicillinresistant Staphylococcus aureus (MRSA) on the surface of burn wounds [14]. Reports of outbreaks of MRSA and evidence of increasing of persistent MRSA strains in burn units have exist. In addition, reports indicate that burn units and ICUs may act as a reservoir for MRSA type of S. aureus. Despite full compliance with infection control programs, acquisition and transmission of MRSA is a continuous problem in burn units [25].
Moreover, rapid generation of antibiotic resistance and the very few new antimicrobial agents being presented each year are very alarming and troublesome [26]. Therefore, the necessity for an alternative therapeutic method other than antibiotics seems absolutely crucial. The use of bacteriophages could potentially be a practical alternative therapeutic strategy. Compared to the costs of antibiotic treatment of staphylococcal infections and patient hospitalization, the cost of phage therapy could be significantly lower [26]. Because of this feature, lytic phages have caught the attention of researchers and industry as a possible antimicrobial agent that would enable us to cope with antibiotic-resistant bacteria such as MRSA [3]. A few studies are present about the MRSA bacteriophages in Thran, Iran, as well as there is a little available information about these local phages and their therapeutic potential. Therefore, in the current study we carried on a search for isolation of new staphylococcal bacteriophages that would be specifically active against MRSA strains isolates from burnt patients.
At first step, a total of 30 MRSA isolates were collected to be used as hosts. Further characterization was carried out in order to confirm that the bacteria were S. aureus as expected and to ascertain their resistance to meticillin. Bacteriophages screening, evaluation of their lytic abilities and host ranges determination were the next steps. In the present study, we successfully isolated six virulent phages against MRSA from Motahari hospital sewage over the course of 6 months. In various studies, such as our study, wastewater has been used as a phage separation reservoir [27-31]. MH-1 titer was determined to be 1×109 PFU/ml therefore; hospital sewage seems to be a good source for isolation of this phage. This suggests that potentially valuable therapeutic phages can be easily recovered from such sources, and that wastewater may have diverse phage populations that can be used for a wide range of applications. Many phages form transparent plaques that are typical of lytic (virulent) phages, whereas phages with the ability to lysogenize host cells (temperate phages) produce opaque plaques. Some phages produce halo plaques, meaning they have semi-transparent areas around the plaques. Halos are due to the release and subsequent activity of soluble enzymes produced by phage that degrade the cell wall [32]. After 18 h incubation at 37°C, the MH-1 phage produced clear plaques without halo with 0.5-2 mm in diameter which is further indicative of the strict lytic nature of the isolated phage.
Phages can detect bacterial cell surface components including lipopolysaccharide (LPS), peptidoglycan, thioacids, outer membrane proteins, oligosaccharides, capsules, and type IV fimbria for the binding process. The specificity of the interaction between phage surface structures and host cell surface receptors is more influential on the bacterial host phage range [33-37]. MH-1 showed broad lytic activities with the tested MRSA and MSSA strains collected from burnt patient. So that most of our S. aureus strains (53 of 60 strains) were lysed by the isolated phage but no plaque production was observed in the examined S. epidermidis, E. faecalis and E. fasium strains. This result suggests that isolated phage was completely specific for S. aureus but there was no correlation found for susceptibility to isolated phage between Meticillin resistance and Methicillin sensitive strains. So MH-1 phage can be used as a disinfectant for circulating S. aureus strains in burn care units and prevent the spread of the bacteria and antibiotic resistance.
Morphological characteristics of phage can be used for their classification. While there is variety of different morphological phage types, most S. aureus phages possess an icosahedral capsid whit double-stranded (ds) DNA as a genome and belong to the order Caudovirales (tailed phages). This order further classified based on the tail morphology into three major families: Podoviridae (characterized by short non-contractile tails), Myoviridae (equipped with long contractile tails) or Siphoviridae (have long, flexible non-contractile tails) [38,39]. Electron microscopy observation of phage MH-1 revealed that this phage belongs to the Myoviridae family. Phages from the Myoviridae family have a double-layered contractile tails composed of an inner tube that covered by an outer sheath and ended by a baseplate (Figure 2). The outer sheath contraction of the tail pushes the tube through the bacterial cell wall and creating a channel for the viral genome delivery into the host cell’s cytoplasm [40]. There are reports of other studies that have isolated myophages for Staphylococcus aureus [41,42].
The one-step growth curve was determined to understand growth of the phage on S. aureus ATCC 43300 as a host. The isolated phage had a short latent period (20 minutes) and a good burst size (190 PFU/cell). The short latent time showed that the time needed to replicate the virus inside the host is very short, and a new generation of phage will be propagate after 20 min. This feature can be considered as a high therapeutic potential for this phage. Also, the high-brush size of this phage could be of relevant interest because it provides the high concentrations needed for phage therapy with little propagation. All of these properties make the isolated phage a suitable candidate for biocontrol of this resistant bacterium.
Digestion of DNA with DNase I, but not with RNase A, is evidence that the isolated phage had the ds DNA as it has been expected to have. To confirm this result, the phage extracted genome was digested with 6 restriction enzymes. The isolated phage DNA samples were sensitive to EcoRV, EcoRI, XbaI, and HaeIII, and exhibited different restriction endonuclease patterns. Additionally, it seems that genomic DNA of isolated phage lacked target sequences recognized by SmaI restriction enzyme. Phages have developed different anti-restriction strategies against restriction modification systems (R-M) of bacteria. This system operates as a defense against phage infections, by means of an endonuclease and methyltransferase enzymes. R-M systems protect methylated bacterial DNA and cut off the foreign unmethylated DNA in the identical sequence. If the recognition oligonucleotide sequences, is present, unmodified DNA molecule will hydrolyse by restriction endonucleases. Point mutations or acquisition of the cognate methylase gene are the strategies which used by bacteriophages to change endonuclease recognition sequences in their genomes [43]. However, for exact determination of recognition sequences in the DNA of phage MH-1, complete genome sequencing is required. In this study, phage MH-1 was isolated and characterized as a new biological strategy to prevent MRSA infection in burnt patient. Its specificity, remarkable lytic effect and broad host-range for MRSA strains emphasized that it has a considerable potential to be used for prophylaxis and treatment of staphylococcal infections. Because of these features MH-1 phage can also be used as a component of anti-staphylococcal or other antibacterial cocktails. It is obvious that additional studies are required to identify the lytic effect of this phage on MRSA isolates from different sources. Additionally, in vivo studies either on experimentally induced infection in animal model as well as in human infection should be carried out in order to fully ascertain the phage therapeutic potentials.
Conclusion
Infection with multi-drug resistant Staphylococcus aureus especially in the burnt patients is a therapeutic problem. In addition, given the cost of mortality and morbidity due to the widespread presence of multidrug-resistant S. aureus strains and the lack of an effective solution to this problem, evaluation of phage therapeutic potentials in human disease management associated with S. aureus seem to be a reasonable proposition. As a result, phage therapy could be an alternative to antibiotics and replace them when they fail. This treatment may help prevent potentially fatal infections in the hospital setting. Moreover, the substantially lower cost of phage therapy is another important reason for its broader consideration in the current period of the global crisis in antibiotic resistance and the health care economy.
Acknowledgment
This study was financially supported by a research grant (No. 26981-30-04-94) for Ph.D. thesis in Iran University of Medical Sciences (Tehran, Iran), for which we are very grateful.
24814
References
- Sussman M (2016) Phage Therapy as a MRSA Treatment. The Science Journal of the Lander College of Arts and Sciences 10: 8.
- Roach DR, Debarbieux L (2017) Phage therapy: awakening a sleeping giant. Emerg Top Life Sci 21: 93-103.
- Abatángelo V, Bacci NP, Boncompain CA, Amadio AA, Carrasco S, et al. (2017) Broad-range lytic bacteriophages that kill Staphylococcus aureus local field strains. PloS one 25 12: e0181671.
- Gutiérrez D, Fernández L, Rodríguez A, García P (2018) Are Phage Lytic Proteins the Secret Weapon To Kill Staphylococcus aureus?. mBio 9: e01923-17.
- Strauß L, Stegger M, Akpaka PE, Alabi A, Breurec S, et al. (2017) Origin, evolution, and global transmission of community-acquired Staphylococcus aureus ST8. Proc Natl Acad Sci USA 20: 201702472.
- Hassoun A, Linden PK, Friedman B (2017) Incidence, prevalence, and management of MRSA bacteremia across patient populations—a review of recent developments in MRSA management and treatment. Critic Care 21: 211.
- Batabyal B, Kundu GK, Biswas S (2012) Methicillin-resistant Staphylococcus aureus: a brief review. Int Res J Biol Sci 1: 65-71.
- Synnott AJ, Kuang Y, Kurimoto M, Yamamichi K, Iwano H, et al. (2009) Isolation from sewage influent and characterization of novel Staphylococcus aureus bacteriophages with wide host ranges and potent lytic capabilities. Appl Environ Microbiol 75: 4483-4490.
- Estrella LA, Quinones J, Henry M, Hannah RM, Pope RK, et al. (2016) Characterization of novel Staphylococcus aureus lytic phage and defining their combinatorial virulence using the OmniLog® system. Bacteriophage 6: e1219440.
- Pincus NB, Reckhow JD, Saleem D, Jammeh ML, Datta SK, et al. (2015) Strain specific phage treatment for Staphylococcus aureus infection is influenced by host immunity and site of infection. PLoS One 10: e0124280.
- Takemura-Uchiyama I, Uchiyama J, Kato SI, Inoue T, Ujihara T, et al. (2013) Evaluating efficacy of bacteriophage therapy against Staphylococcus aureusinfections using a silkworm larval infection model. FEMS microbiology letters 347: 52-60.
- Now MM, Common BS (2012) Skin and soft-tissue infections: classifying and treating a spectrum. CCJMEL 79: 57.
- Merlino JI, Malangoni MA (2007) Complicated skin and soft-tissue infections: diagnostic approach and empiric treatment options. CCJMEL 74: S21.
- Shahsavan S, Emaneini M, Khoshgnab BN, Khoramian B, Asadollahi P, et al.(2012) A high prevalence of mupirocin and macrolide resistance determinant among Staphylococcus aureus strains isolated from burnt patients. Burns 38: 378-382.
- Babakir-Mina M, Othman N, Najmuldeen H, Noori C, Fatah C, et al. (2012) Antibiotic susceptibility of vancomyin and nitrofurantoin in Staphylococcus aureus isolated from burnt patients in Sulaimaniyah, Iraqi Kurdistan. New Microbiologica 35: 439-446.Weber-Dąbrowska B, Górski A (2007) Phage therapy of staphylococcal infections (including MRSA) may be less expensive than antibiotic treatment. PostepyHig Med Dosw (online) 61: 461-465.
- Wang Z, Kong L, Liu Y, Fu Q, Cui Z, et al. (2018) A cell-penetrating peptide fused phage lysin kills intracellular MRSA in keratinocytes and treatment for skin infections of mice. Appl Environ Microbiol 00380
- Iwano H, Inoue Y, Takasago T, Kobayashi H, Furusawa T, et al. (2018) Bacteriophage ΦSA012 Has a Broad Host Range against Staphylococcus aureus and Effective Lytic Capacity in a Mouse Mastitis Model. Biology 7: 8.
- Mohammed-Ali MN, Jamalludeen NM (2015) Isolation and characterization of bacteriophage against methicillin resistant Staphylococcus aureus. J Med Microb Diagn 5: 213.
- Verstappen KM, Tulinski P, Duim B, Fluit AC, Carney J, et al. (2016) The effectiveness of bacteriophages against methicillin-resistant staphylococcus aureus ST398 nasal colonization in pigs. PloS one 11: e0160242
- Clinical Laboratory Standards Institute (2014) Performance Standards for Antimicrobial Susceptibility Testing: Eighteenth Informational Supplement; CLSI Document M100-S18; Clinical Laboratory Standards Institute: Wayne, PA, USA.
- Hudzicki J (2016) Kirby-Bauer Disk Diffusion Susceptibility Test Protocol. 2009. ASM MicrobeLibrary. American Society for Microbiology USA. 2: 3.
- Adriaenssens EM, Wittmann J, Kuhn JH, Turner D, Sullivan MB, et al. (2018) Taxonomy of prokaryotic viruses: 2017 update from the ICTV Bacterial and Archaeal Viruses Subcommittee. Arch Virol 163: 1125-1129.
- Kropinski AM, Clokie MRJ (2015) Bacteriophage methods and protocols. Mol Appl Aspec 2: 565-571.
- Issler-Fisher AC, McKew G, Fisher OM, Harish V, Gottlieb T, et al. (2015) Risk factors for, and the effect of MRSA colonization on the clinical outcomes of severely burnt patients. Burns 41: 1212-1220.
- Kaźmierczak Z, Górski A, Dąbrowska K (2014) Facing antibiotic resistance: Staphylococcus aureus phages as a medical tool. Viruses 6: 2551-2570.
- Sundar MM, Nagananda GS, Das A, Bhattacharya S, Suryan S (2009) Isolation of host-specific bacteriophages from sewage against human pathogens. Asian J Biotechnol 1: 163-170.
- Mattila S, Ruotsalainen P, Jalasvuori M (2015) On-demand isolation of bacteriophages against drug-resistant bacteria for personalized phage therapy. Front Microbiol. 6: 1271.
- Bibi Z, Abbas Z, Rehman SU (2016) The phage P. E1 isolated from hospital sewage reduces the growth of Escherichia coli. Biocontrol Sci Technol 26: 181-188.
- Gautam B, Aryal L, Adhikari S, Rana M, Rajbhanshi A, et al. (2018) Isolation of bacteriophage from Guheswori sewage treatment plant capable of infecting pathogens. Res Pharm Sci 4: 465-470.
- Daneshgar F, SoltanDallal MM, Hosseini F (2018) Isolation of Yersinia enterocolitica Bacteriophage From Hospital Wastewater. Int J Enteric Pathog 6: 18-21.
- Jurczak-Kurek A, Gąsior T, Nejman-Faleńczyk B, Bloch S, Dydecka A, et al. (2016) Biodiversity of bacteriophages: morphological and biological properties of a large group of phages isolated from urban sewage. Sci Rep 6: 34338.
- Abedon S (2006) Phage ecology. In: Calendar R, Abedon ST, editors. The bacteriophages. New York: Oxford University Press pp: 37–46
- Drulis-Kawa Z, Majkowska-Skrobek G, Maciejewska B, Delattre AS, Lavigne R (2012) Learning from bacteriophages-advantages and limitations of phage and phage-encoded protein applications. Curr Protein Pept Sci 13: 699-722.
- Kutter E, Sulakvelidze A (2004) Bacteriophages: biology and applications. CRC Press.
- Sulakvelidze A, Alavidze Z, Morris JG (2001) Bacteriophage therapy. Antimicrob Agents Chemother 45: 649-659.
- Weinbauer MG (2004) Ecology of prokaryotic viruses. FEMS Microbiol Reviews 28: 127-181.
- Xia G, Wolz C (2014) Phages of Staphylococcus aureus and their impact on host evolution. Infect Genet Evol 21: 593-601.
- Fu X, Walter MH, Paredes A, Morais MC, Liu J (2011) The mechanism of DNA ejection in the Bacillus anthracis spore-binding phage 8a revealed by cryo-electron tomography. Virology 421: 141-148.
- Aksyuk AA, Kurochkina LP, Fokine A, Forouhar F, Mesyanzhinov VV, et al. (2011) Structural conservation of the myoviridae phage tail sheath protein fold. Structure 19: 1885-1894.
- El Haddad L, Abdallah NB, Plante PL, Dumaresq J, Katsarava R, et al. (2014) Improving the safety of Staphylococcus aureus polyvalent phages by their production on a Staphylococcus xylosus strain. PLoS One 9: e102600.
- Abatángelo V, Bacci NP, Boncompain CA, Amadio AA, Carrasco S, et al. (2017) Broad-range lytic bacteriophages that kill Staphylococcus aureus local field strains. PloS one 12: e0181671.
- Marek A, Pyzik E, Stępień-Pyśniak D, Urban-Chmiel R, Nowaczek A (2018) Characterization of bacteriophages and their carriage in Staphylococcus aureus isolated from broilers in Poland. British Poultry Science 22:1-8.










