| Introduction |
| Toll-like receptors (TLRs) are key components of the innate immune system, which play role in the recognition of specific microbial components in certain groups of microbes and activate adaptive immunity [1]. For example, TLR2 recognizes bacterial lipoproteins and lipoteichoic acid, TLR3 recognizes viral doublestranded RNA, TLR4 recognizes lipopolysaccharides and TLR5 is the receptor for flagellin [2,3]. |
| Flagellin is the major constituent of bacterial flagella and a virulence factor for Gram-negative and Gram-positive bacteria. The structure of bacterial flagellins is highly conserved and hence TLR5 can detect a wide array of flagellated bacteria including Legionella, Listeria, Pseudomonas and Salmonella [2,3]. Flagellins are expressed by bacteria, particularly pathogenic bacteria, in the lungs and gut and activate epithelial TLR5 signaling. The presence of flagellin thus gives an alarm signal indicating subepithelial invasion and/or disruption of the epithelial barrier function [3]. Upon binding to TLR5 receptor, flagellin triggers activation of MyD88 adaptor protein-dependent signaling pathway and nuclear translocation of NF-kB. Further, MAP kinases are activated, which ultimately induces the maturation of antigenpresenting cells and secretion of proinflammatory chemokines and cytokines, such as tumor necrosis factor-α, necessary for the development of effective immunity [2,4-9]. TLR5 can generate a proinflammatory signal as a homodimer, which suggests it to be the only TLR participating in flagellin recognition [2]. However, TLR5 may require the presence of an adaptor molecule or a coreceptor for efficient ligand recognition and/or signaling [9]. TLR5 is expressed in various cell types including epithelial cells, monocytes, macrophages, dendritic cells, endothelial cells, T-cells and mast cells [10-16]. TLR5 signaling also contributes to mucosal immunity [3,17-19]. TLR5 dysfunction is related to varied human disorders [11,20-24]. |
| Currently, the information about TLR5 signalling is in its infancy. Moreover, accurately locating epitopes is of critical importance for vaccine development, immunodiagnostic tests and antibody production. The techniques presently used for the isolation of TLR5 receptor from crude cell lysate, for example, western blotting, immunoprecipitation, immunohistochemistry and immunocytochemistry, etc. require specific antibodies. The commercial TLR5-specific antibodies (monoclonal or polyclonal) available today are usually produced from host organisms (mouse, rabbit) using large synthetic peptides lacking significant specificity to human TLR5. This non-specificity could possibly be due to the highly conserved nature of TLR5 protein among various mammalian species. The process of antibody production from host organism specific for human TLR5 is thus complex and uneconomical. Thus, in an effort to overcome these problems, in silico predictions have been made about a suitable epitope with optimal length from TLR5 protein sequence, which would be able to generate immune response upon injection into a suitable organism in a highly efficient and economical manner. |
| Antigens are very large and do not bind to a receptor as a whole. Only specific segments, called epitopes bind with a specific antibody [25-27]. Antigenic sites are the amino acid residues of a native protein that are bound by antibodies raised to a native protein or protein fragments or synthetic peptides. Thus, antigenic sites should exhibit at least certain characteristics, which include linearity, high propensity of β-turn, antigenicity, hydrophilicity, flexibility and surface accessibility of amino acid residues [26,28]. Various epitope prediction methods and software are now available [29]. As compared to experimental methods, in silico methods have high potential for efficient and large-scale epitope prediction for antigens at a much reduced cost. |
| For processes involving protein denaturation with a consequent loss of 3-D structure, for example, immunodetection, antibodies recognizing linear epitopes rather than conformational epitopes are selected. The best single method for predicting linear B-cell epitopes is the Hidden Markov Model (HMM). Combining HMM is one of the best propensity scale methods, BepiPred method [30]. This method when tested on the validation data set performed significantly better than other methods examined. Early in silico prediction methods have assumed that there is a good and simple correlation between linear epitope residues and certain propensities. These propensities were hydrophilicity [31,32], flexibility [33], protrusion index (PI) [34], antigenic propensity [35], amino acid pair [36] and β-turns [37]. |
| β-turns are the most common type of non-repetitive structures, which constitute on average 25% of the residues in all protein chains. The formation of β-turns plays an important role in molecular recognition processes. A high degree of correlation between reactivity of anti-peptide antibodies and the tendency for a sequence to form a β-turn has been reported [38]. The formation of β-turns is essential for high-affinity binding to SH2- domains. β-turns are often surface accessible and generally hydrophilic, the two important characteristics of antigenic regions. For this reason these are suitable candidates for being involved in molecular recognition processes. Several researchers have reported that the parts of a protein, which can induce protein-reactive anti-peptide antibodies, mostly reside in regions that have a high tendency to form β-turns [39,40]. |
| Hydrophilicity parameters have been extensively used in algorithms to predict the antigenic amino acid residues [32] presented hydrophilicity HPLC parameters, which correlated best with antigenicity. The correlation of three parameters, namely, hydrophilic, accessible, or mobile regions with the known antigenic sites from immunological studies and/or X-ray crystallographic data for several proteins further improved the prediction of antigenicity. These hydrophilicity parameters [31] algorithm for the prediction of areas of a protein present on the surface. |
Methods
|
|
Sequence retrieval
|
| The protein sequence of human TLR5 receptor protein was retrieved from Uniprot database at https://www.uniprot.org/ and FASTA format of the sequence ID O60602 was obtained (Figure 1). |
|
In silico prediction of TLR5 receptor epitope
|
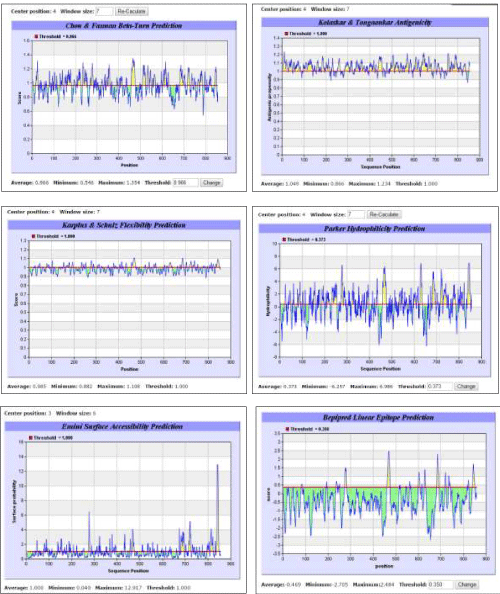
| To predict an epitope from a given sequence of amino acid residues, certain parameters such as probability of having β-turn, antigenicity, surface accessibility of amino acid residues, flexibility, hydrophilicity and linearity, must be ascertained. Pertaining tools for B-cell epitope prediction from Immune Epitope Database (IEDB) were used. All prediction calculations were based on propensity scales for each of the 20 amino acids. |
| Immunoinformatics tool BepiPred developed by Larsen et al. [30] was used for linear epitope prediction. The residues with scores above the threshold were predicted to be part of an epitope. The prediction and analysis of β-turn was done using a mathematical method developed by Chou and Fasman [41] based on the method of Pellequer et al. [37]. Antigenicity was determined on the basis of semi-empirical method developed by Kolaskar and Tongaonkar [35] for the prediction of antigenic determinants on protein antigens. This method is based on physicochemical properties of amino acid residues and their frequencies of occurrence in experimentally known segmental epitopes. This method has been reported by several researchers to predict antigenic determinants with ?75% accuracy. According to this method, hydrophobic residues Cys, Leu and Val, if present on the surface of a protein, are more expected to be a part of antigenic sites. Hydrophilicity was predicted according to the method developed by Parker et al. [32], in which the construction of hydrophilic scale was based on peptide retention times during HPLC on a reversed-phase column. Mathematical model developed by Emini et al. [42] was followed for predicting surface accessibility, wherein a surface residue should have >20 A? of water-accessible surface. The calculation was based on surface accessibility scale on a product. Flexibility was predicted using Karplus and Schulz [33] method. More sophisticated methods for epitope prediction, machine learning methods, Recurrent Neural Network, Support Vector Machine [36,43,44] may be applied to improve performance for linear epitope prediction. |
|
Prediction of host organism for antibody production against human TLR5
|
| Once a peptide sequence was predicted based on all the ideal properties of being an epitope and small size, next step was toa suitable host organism which could be used to initiate an immunogenic reaction upon injecting the predicted peptides, consequently producing antibodies specific for human TLR5. For this purpose predicted epitope sequence of human TLR5 was compared with other host organisms using BLASTp. Mus musculus (NP_058624.2), Gallus gallus (AFV92631.1) and Cirrhinus mrigala (AEQ92867.1) were selected for comparison. |
Results and Discussion
|
| A suitable epitope from the human TLR5 sequence was predicted using the tools for B-cell epitope prediction from IEDB. The prediction methods for β-turn, antigenicity, flexibility, hydrophilicity, surface accessibility of amino acids and linearity revealed a common 16 amino acid sequence with ideal properties for acting as a human TLR5 epitope (Figure 2). Based on recurrence of the same 16 amino acid sequence in all the prediction methods, the peptide predicted to exhibit ideal properties of being an epitope was FSSCSGDQTPSENPSL that occupied a position from 460 to 475. Moreover, this 16 amino acid peptide sequence was unique for TLR5 only. |
| The BLASTp results indicated that TLR5s from two organisms, namely Gallus gallus and Cirrhinus mrigala, showed no significant similarity with the predicted human TLR5 epitope. Comparison of predicted human TLR5 epitope with mouse TLR5, however, showed 69% similarity. To our knowledge, this is the first report indicating variations in TLR5s from human and that from Gallus gallus and Cirrhinus mrigala. Further, it is suggested that the two host organisms, Gallus gallus and Cirrhinus mrigala, might recognize the predicted peptide as a foreign agent and an immunogenic reaction would probably occur to produce antibodies against it. Thus, these two organisms might serve as suitable hosts for large-scale production of human TLR5-specific antibodies. Moreover, as the predicted amino acid peptide sequence was not present in other forms of TLRs, any response invoked in host organisms would not be due to any other form of TLR. Further advantage associated with these organisms is their ease of availability from poultry and fisheries, respectively, for the purpose of experimental validation. The results of the present study suggest a probability of commercial success for providing valuable source of antibody against human TLR5 for application in research in time- and cost-effective manner. |
Figures at a glance
|
 |
 |
| Figure 1 |
Figure 2 |
|
| |







